- Record: found
- Abstract: found
- Article: found
A secretomic view of woody and nonwoody lignocellulose degradation by Pleurotus ostreatus
Read this article at
Abstract
Background
Pleurotus ostreatus is the second edible mushroom worldwide, and a model fungus for delignification applications, with the advantage of growing on woody and nonwoody feedstocks. Its sequenced genome is available, and this gave us the opportunity to perform proteomic studies to identify the enzymes overproduced in lignocellulose cultures.
Results
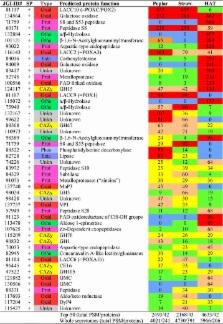
Monokaryotic P. ostreatus (PC9) was grown with poplar wood or wheat straw as the sole C/N source and the extracellular proteins were analyzed, together with those from glucose medium. Using nano-liquid chromatography coupled to tandem mass spectrometry of whole-protein hydrolyzate, over five-hundred proteins were identified. Thirty-four percent were unique of the straw cultures, while only 15 and 6 % were unique of the glucose and poplar cultures, respectively (20 % were produced under the three conditions, and additional 19 % were shared by the two lignocellulose cultures). Semi-quantitative analysis showed oxidoreductases as the main protein type both in the poplar (39 % total abundance) and straw (31 %) secretomes, while carbohydrate-active enzymes (CAZys) were only slightly overproduced (14–16 %). Laccase 10 (LACC10) was the main protein in the two lignocellulose secretomes (10–14 %) and, together with LACC2, LACC9, LACC6, versatile peroxidase 1 (VP1), and manganese peroxidase 3 (MnP3), were strongly overproduced in the lignocellulose cultures. Seven CAZys were also among the top-50 proteins, but only CE16 acetylesterase was overproduced on lignocellulose. When the woody and nonwoody secretomes were compared, GH1 and GH3 β-glycosidases were more abundant on poplar and straw, respectively and, among less abundant proteins, VP2 was overproduced on straw, while VP3 was only found on poplar. The treated lignocellulosic substrates were analyzed by two-dimensional nuclear magnetic resonance (2D NMR), and a decrease of lignin relative to carbohydrate signals was observed, together with the disappearance of some minor lignin substructures, and an increase of sugar reducing ends.
Conclusions
Oxidoreductases are strongly induced when P. ostreatus grows on woody and nonwoody lignocellulosic substrates. One laccase occupied the first position in both secretomes, and three more were overproduced together with one VP and one MnP, suggesting an important role in lignocellulose degradation. Preferential removal of lignin vs carbohydrates was shown by 2D NMR, in agreement with the above secretomic results.
Related collections
Most cited references89

- Record: found
- Abstract: found
- Article: found
Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes
- Record: found
- Abstract: found
- Article: not found
Extensive sampling of basidiomycete genomes demonstrates inadequacy of the white-rot/brown-rot paradigm for wood decay fungi.
- Record: found
- Abstract: found
- Article: not found