- Record: found
- Abstract: found
- Article: found
Reorganization of a synthetic microbial consortium for one-step vitamin C fermentation
Read this article at
Abstract
Background
In the industry, the conventional two-step fermentation method was used to produce 2-keto- l-gulonic acid (2-KGA), the precursor of vitamin C, by three strains, namely, Gluconobacter oxydans, Bacillus spp. and Ketogulonicigenium vulgare. Despite its high production efficiency, the long incubation period and an additional second sterilization process inhibit the further development. Therefore, we aimed to reorganize a synthetic consortium of G. oxydans and K. vulgare for one-step fermentation of 2-KGA and enhance the symbiotic interaction between microorganisms to perform better.
Results
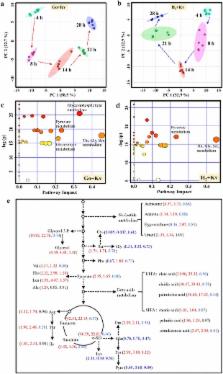
During the fermentation, competition for sorbose of G. oxydans arose when co-cultured with K. vulgare. In this study, the competition between the two microbes was alleviated and their mutualism was enhanced by deleting genes involved in sorbose metabolism of G. oxydans. In the engineered synthetic consortium (H 6 + Kv), the yield of 2-KGA (mol/mol) against d-sorbitol reached 89.7 % within 36 h, increased by 29.6 %. Furthermore, metabolomic analysis was used to verify the enhancement of the symbiotic relationship and to provide us potential strategies for improving the synthetic consortium. Additionally, a significant redistribution of metabolism occurred by co-culturing the K. vulgare with the engineered G. oxydans, mainly reflected in the increased TCA cycle, purine, and fatty acid metabolism.
Conclusions
We reorganized and optimized a synthetic consortium of G. oxydans and K. vulgare to produce 2-KGA directly from d-sorbitol. The yield of 2-KGA was comparable to that of the conventional two-step fermentation. The metabolic interaction between the strains was further investigated by metabolomics, which verified the enhancement of the mutualism between the microbes and gave us a better understanding of the synthetic consortium.
Related collections
Most cited references21
- Record: found
- Abstract: found
- Article: not found
Natural strategies for the spatial optimization of metabolism in synthetic biology.
- Record: found
- Abstract: found
- Article: not found
Microbial metabolic exchange--the chemotype-to-phenotype link.
- Record: found
- Abstract: found
- Article: not found