- Record: found
- Abstract: found
- Article: not found
Machine Learning-Assisted Real-Time Polymerase Chain Reaction and High-Resolution Melt Analysis for SARS-CoV-2 Variant Identification

Read this article at
Abstract
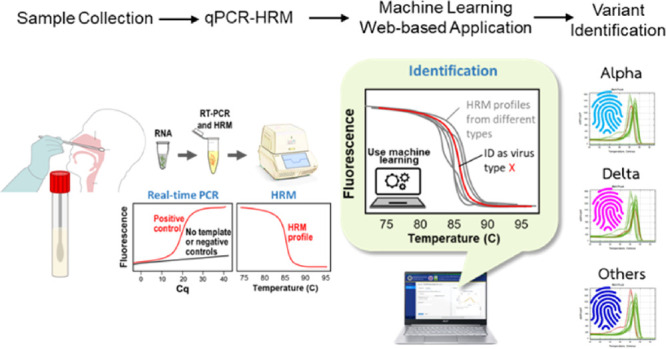
Since the declaration of COVID-19 as a pandemic in early 2020, multiple variants of the severe acute respiratory syndrome-related coronavirus (SARS-CoV-2) have been detected. The emergence of multiple variants has raised concerns due to their impact on public health. Therefore, it is crucial to distinguish between different viral variants. Here, we developed a machine learning web-based application for SARS-CoV-2 variant identification via duplex real-time polymerase chain reaction (PCR) coupled with high-resolution melt (qPCR-HRM) analysis. As a proof-of-concept, we investigated the platform’s ability to identify the Alpha, Delta, and wild-type strains using two sets of primers. The duplex qPCR-HRM could identify the two variants reliably in as low as 100 copies/μL. Finally, the platform was validated with 167 nasopharyngeal swab samples, which gave a sensitivity of 95.2%. This work demonstrates the potential for use as automated, cost-effective, and large-scale viral variant surveillance.
Related collections
Most cited references45
- Record: found
- Abstract: found
- Article: found
Neutralising antibody activity against SARS-CoV-2 VOCs B.1.617.2 and B.1.351 by BNT162b2 vaccination

- Record: found
- Abstract: found
- Article: not found
