- Record: found
- Abstract: found
- Article: found
Structural Insights for Activation of Retinal Guanylate Cyclase by GCAP1
Read this article at
Abstract
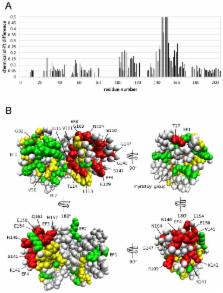
Guanylyl cyclase activating protein 1 (GCAP1), a member of the neuronal calcium sensor (NCS) subclass of the calmodulin superfamily, confers Ca 2+-sensitive activation of retinal guanylyl cyclase 1 (RetGC1) upon light activation of photoreceptor cells. Here we present NMR assignments and functional analysis to probe Ca 2+-dependent structural changes in GCAP1 that control activation of RetGC. NMR assignments were obtained for both the Ca 2+-saturated inhibitory state of GCAP1 versus a GCAP1 mutant (D144N/D148G, called EF4mut), which lacks Ca 2+ binding in EF-hand 4 and models the Ca 2+-free/Mg 2+-bound activator state of GCAP1. NMR chemical shifts of backbone resonances for Ca 2+-saturated wild type GCAP1 are overall similar to those of EF4mut, suggesting a similar main chain structure for assigned residues in both the Ca 2+-free activator and Ca 2+-bound inhibitor states. This contrasts with large Ca 2+-induced chemical shift differences and hence dramatic structural changes seen for other NCS proteins including recoverin and NCS-1. The largest chemical shift differences between GCAP1 and EF4mut are seen for residues in EF4 (S141, K142, V145, N146, G147, G149, E150, L153, E154, M157, E158, Q161, L166), but mutagenesis of EF4 residues (F140A, K142D, L153R, L166R) had little effect on RetGC1 activation. A few GCAP1 residues in EF-hand 1 (K23, T27, G32) also show large chemical shift differences, and two of the mutations (K23D and G32N) each decrease the activation of RetGC, consistent with a functional conformational change in EF1. GCAP1 residues at the domain interface (V77, A78, L82) have NMR resonances that are exchange broadened, suggesting these residues may be conformationally dynamic, consistent with previous studies showing these residues are in a region essential for activating RetGC1.
Related collections
Most cited references62
- Record: found
- Abstract: found
- Article: not found
Neuronal calcium sensor proteins: generating diversity in neuronal Ca2+ signalling.
- Record: found
- Abstract: found
- Article: not found
Backbone dynamics of Escherichia coli ribonuclease HI: correlations with structure and function in an active enzyme.
- Record: found
- Abstract: found
- Article: not found