- Record: found
- Abstract: found
- Article: found
Epigenetic integrity of paternal imprints enhances the developmental potential of androgenetic haploid embryonic stem cells
Read this article at
Abstract
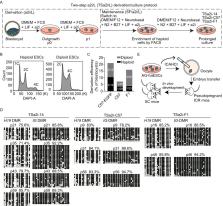
The use of two inhibitors of Mek1/2 and Gsk3β (2i) promotes the generation of mouse diploid and haploid embryonic stem cells (ESCs) from the inner cell mass of biparental and uniparental blastocysts, respectively. However, a system enabling long-term maintenance of imprints in ESCs has proven challenging. Here, we report that the use of a two-step a2i (alternative two inhibitors of Src and Gsk3β, TSa2i) derivation/culture protocol results in the establishment of androgenetic haploid ESCs (AG-haESCs) with stable DNA methylation at paternal DMRs (differentially DNA methylated regions) up to passage 60 that can efficiently support generating mice upon oocyte injection. We also show coexistence of H3K9me3 marks and ZFP57 bindings with intact DMR methylations. Furthermore, we demonstrate that TSa2i-treated AG-haESCs are a heterogeneous cell population regarding paternal DMR methylation. Strikingly, AG-haESCs with late passages display increased paternal-DMR methylations and improved developmental potential compared to early-passage cells, in part through the enhanced proliferation of H19-DMR hypermethylated cells. Together, we establish AG-haESCs that can long-term maintain paternal imprints.
Related collections
Most cited references52
- Record: found
- Abstract: not found
- Article: not found
Establishment in culture of pluripotential cells from mouse embryos.
- Record: found
- Abstract: found
- Article: not found
DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA.
- Record: found
- Abstract: found
- Article: not found