- Record: found
- Abstract: found
- Article: found
Mycobacterium pseudoshottsii in Mediterranean Fish Farms: New Trouble for European Aquaculture?
Read this article at
Abstract
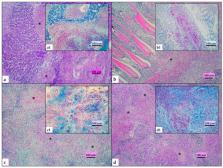
Mycobacterium pseudoshottsii, a slow-growing mycobacterium closely related to M. marinum, has been isolated only in wild fish in the United States and in Japanese fish farms to date. Here, we report cases of mortality in three farmed fish species ( Dicentrarchus labrax, Sparus aurata, and Sciaenops ocellatus) caused by M. pseudoshottsii in Italy. Samples underwent necropsy, histology, and culture with pathogen identification based on PCR and sequencing of housekeeping genes (16S rRNA, hsp65, rpoB). Multifocal to coalescing granulomatous and necrotizing inflammation with acid-fast bacilli were observed in the parenchymatous organs, from which M. pseudoshottsii was isolated and identified. Phylogenetic analysis confirmed the results of gene sequencing and allowed subdivision of the isolates into three distinct groups. M. pseudoshottsii poses a potential threat for Mediterranean aquaculture. Its origin in the area under study needs to be clarified, as well as the threat to the farmed fish species.
Related collections
Most cited references44
- Record: found
- Abstract: found
- Article: not found
Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis.
- Record: found
- Abstract: found
- Article: not found
rpoB-based identification of nonpigmented and late-pigmenting rapidly growing mycobacteria.
- Record: found
- Abstract: not found
- Article: not found