- Record: found
- Abstract: found
- Article: found
Mechanism and regulation of sorbicillin biosynthesis by Penicillium chrysogenum
Read this article at
Summary
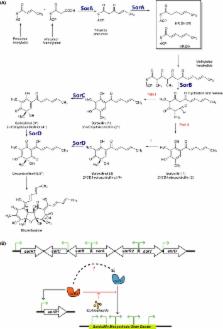
Penicillium chrysogenum is a filamentous fungus that is used to produce β‐lactams at an industrial scale. At an early stage of classical strain improvement, the ability to produce the yellow‐coloured sorbicillinoids was lost through mutation. Sorbicillinoids are highly bioactive of great pharmaceutical interest. By repair of a critical mutation in one of the two polyketide synthases in an industrial P. chrysogenum strain, sorbicillinoid production was restored at high levels. Using this strain, the sorbicillin biosynthesis pathway was elucidated through gene deletion, overexpression and metabolite profiling. The polyketide synthase enzymes SorA and SorB are required to generate the key intermediates sorbicillin and dihydrosorbicillin, which are subsequently converted to (dihydro)sorbillinol by the FAD‐dependent monooxygenase SorC and into the final product oxosorbicillinol by the oxidoreductase SorD. Deletion of either of the two pks genes not only impacted the overall production but also strongly reduce the expression of the pathway genes. Expression is regulated through the interplay of two transcriptional regulators: SorR1 and SorR2. SorR1 acts as a transcriptional activator, while SorR2 controls the expression of sorR1. Furthermore, the sorbicillinoid pathway is regulated through a novel autoinduction mechanism where sorbicillinoids activate transcription.
Related collections
Most cited references31
- Record: found
- Abstract: found
- Article: not found
Regulation of fungal secondary metabolism.
- Record: found
- Abstract: found
- Article: not found
Regulation of secondary metabolism in filamentous fungi.
- Record: found
- Abstract: found
- Article: not found