- Record: found
- Abstract: found
- Article: found
The long non-coding RNA CYTOR drives colorectal cancer progression by interacting with NCL and Sam68
Read this article at
Abstract
Background
Long non-coding RNAs (lncRNAs) function as key molecules in cancer progression. The lncRNA CYTOR plays oncogenic roles in multiple types of cancer, yet the detailed molecular mechanisms of those roles remain unknown. The aim of this study was to investigate the clinical significance, biological function and interacting partners of CYTOR in colorectal cancer (CRC).
Methods
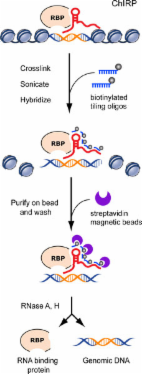
A systematic and comprehensive analysis of CYTOR expression was performed in 138 CRC samples and in the TCGA and GEO databases. Biological function was investigated through knockdown and overexpression of CYTOR in vitro and in vivo. In addition, its protein binding partner was identified and validated using ChIRP-MS and RNA immunoprecipitation assays. Their key interaction sites on CYTOR were verified by CRISPR/Cas9 and a series of mutant constructs. Furthermore, the downstream targets of CYTOR were confirmed via immunoblotting and luciferase reporter assays.
Results
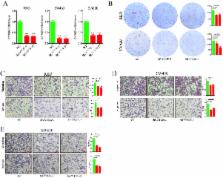
CYTOR was significantly up-regulated in CRC samples and associated with poor prognosis, promoting proliferation and metastasis in vitro and in vivo. NCL and Sam68 could recognize their specific motifs and directly bind to EXON1 of CYTOR. Moreover, EXON1 was the key functional site mediating the interaction of CYTOR with NCL and Sam68. NCL and Sam68 functioned as oncogenes to promote CRC progression. Furthermore, we confirmed that the heterotrimeric complex of CYTOR, NCL and Sam68 activated the NF-κB pathway and EMT to contribute to CRC progression.
Related collections
Most cited references25
- Record: found
- Abstract: found
- Article: not found
Stem cell divisions, somatic mutations, cancer etiology, and cancer prevention
- Record: found
- Abstract: found
- Article: not found
Regulation of transcription by long noncoding RNAs.
- Record: found
- Abstract: found
- Article: found