- Record: found
- Abstract: found
- Article: found
Structural and Functional Studies of Nonstructural Protein 2 of the Hepatitis C Virus Reveal Its Key Role as Organizer of Virion Assembly
Read this article at
Abstract
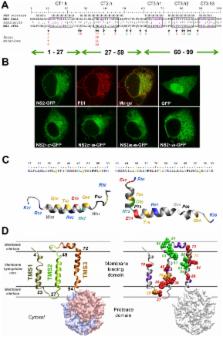
Non-structural protein 2 (NS2) plays an important role in hepatitis C virus (HCV) assembly, but neither the exact contribution of this protein to the assembly process nor its complete structure are known. In this study we used a combination of genetic, biochemical and structural methods to decipher the role of NS2 in infectious virus particle formation. A large panel of NS2 mutations targeting the N-terminal membrane binding region was generated. They were selected based on a membrane topology model that we established by determining the NMR structures of N-terminal NS2 transmembrane segments. Mutants affected in virion assembly, but not RNA replication, were selected for pseudoreversion in cell culture. Rescue mutations restoring virus assembly to various degrees emerged in E2, p7, NS3 and NS2 itself arguing for an interaction between these proteins. To confirm this assumption we developed a fully functional JFH1 genome expressing an N-terminally tagged NS2 demonstrating efficient pull-down of NS2 with p7, E2 and NS3 and, to a lower extent, NS5A. Several of the mutations blocking virus assembly disrupted some of these interactions that were restored to various degrees by those pseudoreversions that also restored assembly. Immunofluorescence analyses revealed a time-dependent NS2 colocalization with E2 at sites close to lipid droplets (LDs) together with NS3 and NS5A. Importantly, NS2 of a mutant defective in assembly abrogates NS2 colocalization around LDs with E2 and NS3, which is restored by a pseudoreversion in p7, whereas NS5A is recruited to LDs in an NS2-independent manner. In conclusion, our results suggest that NS2 orchestrates HCV particle formation by participation in multiple protein-protein interactions required for their recruitment to assembly sites in close proximity of LDs.
Author Summary
Formation of infectious virus particles (assembly) is a complex process by which structural proteins and the viral genome must be transferred to the same subcellular sites to allow their direct or indirect interaction. In case of the hepatitis C virus (HCV), this process appears to take place in close proximity of lipid droplets (LDs) and requires in addition to the structural proteins core, envelope glycoprotein 1 (E1) and E2 two auxiliary factors, designated p7 and nonstructural protein 2 (NS2), contributing to virion formation by unknown mechanisms. In this study we used a combination of structural, genetic and biochemical assays to study the role of NS2 in HCV assembly. By using nuclear magnetic resonance spectroscopy of NS2 peptides we established a membrane topology model of the amino-terminal membrane binding domain of NS2. We found that this protein participates in multiple interactions with E2, p7, NS3 and NS5A that appear to recruit the viral proteins to sites in close proximity of LDs. In this respect, NS2 is a key organizer of the assembly of infectious HCV particles.
Related collections
Most cited references55
- Record: found
- Abstract: found
- Article: not found
The lipid droplet is an important organelle for hepatitis C virus production.
- Record: found
- Abstract: found
- Article: not found
Construction and characterization of infectious intragenotypic and intergenotypic hepatitis C virus chimeras.
- Record: found
- Abstract: found
- Article: not found