- Record: found
- Abstract: found
- Article: found
Molecular characterization of adenocarcinomas arising in the urinary bladder following augmentation cystoplasty: a multi-institutional study✩

Read this article at
Summary
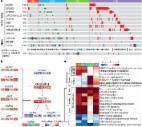
Development of malignancy is a rare complication following augmentation cystoplasty, and the majority of tumors observed in this setting are adenocarcinomas. Here, we sought to genetically profile these tumors by targeted DNA sequencing of a multi-institutional cohort of adenocarcinomas that developed in the urinary bladder following augmentation cystoplasty. Carcinomas arising in the urinary bladder of patients with history of augmentation cystoplasty were obtained from 4 major academic institutions, with cases diagnosed as urothelial carcinoma excluded from the study. The cases were analyzed using a DNA sequencing panel that includes 529 genes and genome-wide copy number assessment. The most frequently altered genes included TP53, KRAS, and MYC, and the vast majority of cases demonstrated mutational profiles consistent with gastrointestinal adenocarcinomas. One case demonstrated an EML4::ALK fusion together with an MSH3 frameshift mutation and hypermutated phenotype, characteristic of a rare but aggressive subtype of colorectal adenocarcinoma that may benefit from targeted ALK inhibition therapy. Importantly, six other tumors in the cohort also had potentially targetable molecular alterations, involving ATM (2 cases ), BRCA1 (2 cases), EGFR (1 case), and ERBB2 (1 case). To our knowledge, this study represents the most comprehensive molecular characterization to date of adenocarcinomas arising in the urinary bladder following augmentation cystoplasty. Despite the unique environment of the augmented tissue, the resulting tumors demonstrate a spectrum of driver mutations similar to that of primary gastrointestinal adenocarcinomas. Importantly, molecular alterations potentially amenable to targeted therapy were identified in the majority of cases.
Related collections
Most cited references17

- Record: found
- Abstract: found
- Article: found
Comprehensive Molecular Characterization of Human Colon and Rectal Cancer
- Record: found
- Abstract: found
- Article: found
Comprehensive molecular characterization of gastric adenocarcinoma

- Record: found
- Abstract: found
- Article: found