- Record: found
- Abstract: found
- Article: found
Immunization with SARS Coronavirus Vaccines Leads to Pulmonary Immunopathology on Challenge with the SARS Virus
Read this article at
Abstract
Background
Severe acute respiratory syndrome (SARS) emerged in China in 2002 and spread to other countries before brought under control. Because of a concern for reemergence or a deliberate release of the SARS coronavirus, vaccine development was initiated. Evaluations of an inactivated whole virus vaccine in ferrets and nonhuman primates and a virus-like-particle vaccine in mice induced protection against infection but challenged animals exhibited an immunopathologic-type lung disease.
Design
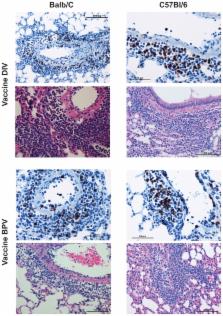
Four candidate vaccines for humans with or without alum adjuvant were evaluated in a mouse model of SARS, a VLP vaccine, the vaccine given to ferrets and NHP, another whole virus vaccine and an rDNA-produced S protein. Balb/c or C57BL/6 mice were vaccinated IM on day 0 and 28 and sacrificed for serum antibody measurements or challenged with live virus on day 56. On day 58, challenged mice were sacrificed and lungs obtained for virus and histopathology.
Results
All vaccines induced serum neutralizing antibody with increasing dosages and/or alum significantly increasing responses. Significant reductions of SARS-CoV two days after challenge was seen for all vaccines and prior live SARS-CoV. All mice exhibited histopathologic changes in lungs two days after challenge including all animals vaccinated (Balb/C and C57BL/6) or given live virus, influenza vaccine, or PBS suggesting infection occurred in all. Histopathology seen in animals given one of the SARS-CoV vaccines was uniformly a Th2-type immunopathology with prominent eosinophil infiltration, confirmed with special eosinophil stains. The pathologic changes seen in all control groups lacked the eosinophil prominence.
Conclusions
These SARS-CoV vaccines all induced antibody and protection against infection with SARS-CoV. However, challenge of mice given any of the vaccines led to occurrence of Th2-type immunopathology suggesting hypersensitivity to SARS-CoV components was induced. Caution in proceeding to application of a SARS-CoV vaccine in humans is indicated.
Related collections
Most cited references55
- Record: found
- Abstract: found
- Article: not found
Identification of a Novel Coronavirus in Patients with Severe Acute Respiratory Syndrome
- Record: found
- Abstract: found
- Article: not found
A novel coronavirus associated with severe acute respiratory syndrome.
- Record: found
- Abstract: found
- Article: not found