- Record: found
- Abstract: found
- Article: not found
Targeted nanopore sequencing by real-time mapping of raw electrical signal with UNCALLED
Read this article at
Abstract
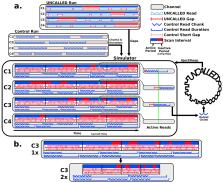
Conventional targeted sequencing methods eliminate many of the benefits of nanopore sequencing, such as the ability to accurately detect structural variants (SVs) or epigenetic modifications. The ReadUntil method allows nanopore devices to selectively eject reads from pores in real-time, which could enable purely computational targeted sequencing. However this requires rapid identification of on-target reads, and most mapping methods require computationally intensive basecalling. We present UNCALLED ( github.com/skovaka/UNCALLED), an open-source mapper that rapidly matches streaming nanopore current signals to a reference sequence. UNCALLED probabilistically considers k-mers that the signal could represent, and then prunes the candidates based on the reference encoded within an FM-index. We used UNCALLED to deplete sequencing of known bacterial genomes within a metagenomics community, enriching the remaining species by 4.46 fold. UNCALLED also enriched 148 human genes associated with hereditary cancers to 29.6x coverage using one MinION flowcell, enabling accurate detection of SNPs, indels, SVs, and methylation in these genes.
Related collections
Most cited references50

- Record: found
- Abstract: found
- Article: found
BEDTools: a flexible suite of utilities for comparing genomic features
- Record: found
- Abstract: found
- Article: not found
HISAT: a fast spliced aligner with low memory requirements.
- Record: found
- Abstract: found
- Article: not found
