- Record: found
- Abstract: found
- Article: found
Rapid turnover of pathogen-blocking Wolbachia and their incompatibility loci
Read this article at
Abstract
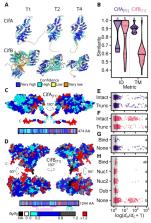
At least half of all insect species carry maternally inherited Wolbachia alphaproteobacteria, making Wolbachia the most common endosymbionts in nature. Wolbachia spread to high frequencies is often due to cytoplasmic incompatibility (CI), a Wolbachia-induced sperm modification that kills embryos without Wolbachia. Several CI-causing Wolbachia variants, including wMel from Drosophila melanogaster, also block viruses. Establishing pathogen-blocking wMel in natural Aedes aegypti mosquito populations has reduced dengue disease incidence, with one study reporting about 85% reduction when wMel frequency is high. However, wMel transinfection establishment is challenging in many environments, highlighting the importance of identifying CI-causing Wolbachia variants that stably persist in diverse hosts and habitats. We demonstrate that wMel-like variants have naturally established in widely distributed holometabolous dipteran and hymenopteran insects that diverged approximately 350 million years ago, with wMel variants spreading rapidly among these hosts over only the last 100,000 years. Wolbachia genomes contain prophages that encode CI-causing operons ( cifs). These cifs move among Wolbachia genomes – with and without prophages – even more rapidly than Wolbachia move among insect hosts. Our results shed light on how rapid host switching and horizontal gene transfer contribute to Wolbachia and cif diversity in nature. The diverse wMel variants we report here from hosts present in different climates offer many new options for broadening Wolbachia-based biocontrol of diseases and pests.
Related collections
Most cited references66
- Record: found
- Abstract: found
- Article: not found
Time dependency of molecular rate estimates and systematic overestimation of recent divergence times.
- Record: found
- Abstract: found
- Article: not found
Successful establishment of Wolbachia in Aedes populations to suppress dengue transmission.
- Record: found
- Abstract: found
- Article: not found