- Record: found
- Abstract: found
- Article: not found
Vaccine Breakthrough Infections with SARS-CoV-2 Variants
Read this article at
Summary
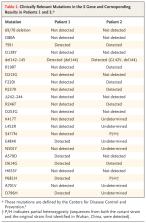
Emerging variants of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) are of clinical concern. In a cohort of 417 persons who had received the second dose of BNT162b2 (Pfizer–BioNTech) or mRNA-1273 (Moderna) vaccine at least 2 weeks previously, we identified 2 women with vaccine breakthrough infection. Despite evidence of vaccine efficacy in both women, symptoms of coronavirus disease 2019 developed, and they tested positive for SARS-CoV-2 by polymerase-chain-reaction testing. Viral sequencing revealed variants of likely clinical importance, including E484K in 1 woman and three mutations (T95I, del142–144, and D614G) in both. These observations indicate a potential risk of illness after successful vaccination and subsequent infection with variant virus, and they provide support for continued efforts to prevent and diagnose infection and to characterize variants in vaccinated persons. (Funded by the National Institutes of Health and others.)
Related collections
Most cited references12
- Record: found
- Abstract: found
- Article: not found
A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology
- Record: found
- Abstract: found
- Article: not found
Convergent Antibody Responses to SARS-CoV-2 in Convalescent Individuals
- Record: found
- Abstract: found
- Article: not found
