- Record: found
- Abstract: found
- Article: found
Denaturation of proteins by surfactants studied by the Taylor dispersion analysis
Read this article at
Abstract
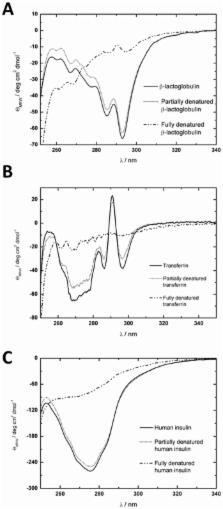
We showed that the Taylor Dispersion Analysis (TDA) is a fast and easy to use method for the study of denaturation proteins. We applied TDA to study denaturation of β-lactoglobulin, transferrin, and human insulin by anionic surfactant sodium dodecyl sulfate (SDS). A series of measurements at constant protein concentration (for transferrin was 1.9 x 10 −5 M, for β- lactoglobulin was 7.6 x 10 −5 M, and for insulin was 1.2 x 10 −4 M) and varying SDS concentrations were carried out in the phosphate-buffered saline (PBS). The structural changes were analyzed based on the diffusion coefficients of the complexes formed at various surfactant concentrations. The concentration of surfactant was varied in the range from 1.2 x 10 −4 M to 8.7 x 10 −2 M. We determined the minimum concentration of the surfactant necessary to change the native conformation of the proteins. The minimal concentration of SDS for β-lactoglobulin and transferrin was 4.3 x 10 −4 M and for insulin 2.3 x 10 −4 M. To evaluate the TDA as a novel method for studying denaturation of proteins we also applied other methods i.e. electronic circular dichroism (ECD) and dynamic light scattering (DLS) to study the same phenomenon. The results obtained using these methods were in agreement with the results from TDA.
Related collections
Most cited references36
- Record: found
- Abstract: found
- Article: not found
Protein unfolding in detergents: effect of micelle structure, ionic strength, pH, and temperature.
- Record: found
- Abstract: found
- Article: not found
Simple detection of protein soft structure changes.
- Record: found
- Abstract: found
- Article: not found