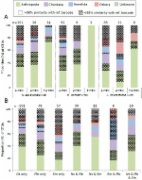
Introduction The PCR-based analysis of homologous genes has become one of the most powerful approaches for species detection and identification, particularly with the recent availability of Next Generation Sequencing platforms (NGS) making it possible to identify species composition from a broad range of environmental samples. Identifying species from these samples relies on the ability to match sequences with reference barcodes for taxonomic identification. Unfortunately, most studies of environmental samples have targeted ribosomal markers, despite the fact that the mitochondrial Cytochrome c Oxidase subunit I gene (COI) is by far the most widely available sequence region in public reference libraries. This is largely because the available versatile (“universal”) COI primers target the 658 barcoding region, whose size is considered too large for many NGS applications. Moreover, traditional barcoding primers are known to be poorly conserved across some taxonomic groups. Results We first design a new PCR primer within the highly variable mitochondrial COI region, the “mlCOIintF” primer. We then show that this newly designed forward primer combined with the “jgHCO2198” reverse primer to target a 313 bp fragment performs well across metazoan diversity, with higher success rates than versatile primer sets traditionally used for DNA barcoding (i.e. LCO1490/HCO2198). Finally, we demonstrate how the shorter COI fragment coupled with an efficient bioinformatics pipeline can be used to characterize species diversity from environmental samples by pyrosequencing. We examine the gut contents of three species of planktivorous and benthivorous coral reef fish (family: Apogonidae and Holocentridae). After the removal of dubious COI sequences, we obtained a total of 334 prey Operational Taxonomic Units (OTUs) belonging to 14 phyla from 16 fish guts. Of these, 52.5% matched a reference barcode (>98% sequence similarity) and an additional 32% could be assigned to a higher taxonomic level using Bayesian assignment. Conclusions The molecular analysis of gut contents targeting the 313 COI fragment using the newly designed mlCOIintF primer in combination with the jgHCO2198 primer offers enormous promise for metazoan metabarcoding studies. We believe that this primer set will be a valuable asset for a range of applications from large-scale biodiversity assessments to food web studies.