- Record: found
- Abstract: found
- Article: found
An Engineered Version of Human PON2 Opens the Way to Understand the Role of Its Post-Translational Modifications in Modulating Catalytic Activity
Read this article at
Abstract
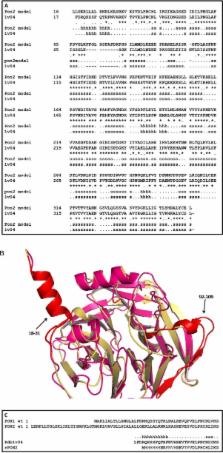
The human paraoxonase 2 (PON2) has been described as a highly specific lactonase hydrolysing the quorum sensing molecule N-(3-oxododecanoyl)-L-homoserine lactone (3OC12-HSL) and having secondary esterase but not phosphotriesterase activity, in contrast with the related enzymes PON1 and PON3. It has been suggested that PON2 enzyme activity is dependent on glycosylation and its N-terminal region has been recently demonstrated to be a transmembrane domain mediating association to membranes. In the present study we describe a mutated form of PON2, lacking the above N-terminal region, which has been further stabilized by the insertion of six amino acidic substitutions. The engineered version, hence forth called rPON2, has been over-expressed in E. coli, refolded from inclusion bodies and purified, yielding an enzyme with the same characteristics as the full length enzyme. Therefore the first conclusion of this work was that the catalytic activity is independent from the N-terminus and protein glycosylation. The kinetic characterization confirmed the primary activity on 3OC12-HSL; accordingly, in vitro experiments of inhibition of the biofilm formed by Pseudomonas aeruginosa (PAO1) have demonstrated that rPON2 is more effective than PON1. In addition, we observed small but significant activity against organophosphorothiotes pesticides, m-parathion, coumaphos and malathion.The availability of fair amount of active protein allowed to pinpoint, by mass-spectrometry, ubiquitination of Lys 168 induced in rPON2 by HeLa extract and to correlate such post-translational modification to the modulation of catalytic activity. A mutational analysis of the modified residue confirmed the result.
Related collections
Most cited references37
- Record: found
- Abstract: found
- Article: not found
Systematic and quantitative assessment of the ubiquitin-modified proteome.
- Record: found
- Abstract: not found
- Article: not found
Use of T7 RNA polymerase to direct expression of cloned genes.
- Record: found
- Abstract: found
- Article: not found