- Record: found
- Abstract: found
- Article: found
A synopsis of the saddle fungi ( Helvella: Ascomycota) in Europe – species delimitation, taxonomy and typification
Read this article at
Abstract
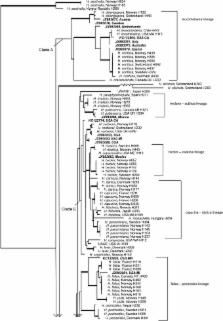
Helvella is a widespread, speciose genus of large apothecial ascomycetes ( Pezizomycete: Pezizales) that are found in terrestrial biomes of the Northern and Southern Hemispheres. This study represents a beginning on assessing species limits and applying correct names for Helvella species based on type material and specimens in the university herbaria (fungaria) of Copenhagen (C), Harvard (FH) and Oslo (O). We use morphology and phylogenetic evidence from four loci – heat shock protein 90 ( hsp), translation elongation factor alpha ( tef), RNA polymerase II ( rpb2) and the nuclear large subunit ribosomal DNA (LSU) – to assess species boundaries in an expanded sample of Helvella specimens from Europe. We combine the morphological and phylogenetic information from 55 Helvella species from Europe with a small sample of Helvella species from other regions of the world. Little intraspecific variation was detected within the species using these molecular markers; hsp and rpb2 markers provided useful barcodes for species delimitation in this genus, while LSU provided more variable resolution among the pertinent species. We discuss typification issues and identify molecular characteristics for 55 European Helvella species, designate neo- and epitypes for 30 species, and describe seven Helvella species new to science, i.e., H. alpicola, H. alpina, H. carnosa, H. danica, H. nannfeldtii, H. pubescens and H. scyphoides.
Related collections
Most cited references104
- Record: found
- Abstract: found
- Article: not found
Phylogenetic species recognition and species concepts in fungi.
- Record: found
- Abstract: found
- Article: not found