- Record: found
- Abstract: found
- Article: not found
Ultrasensitive and Simultaneous Detection of Two Specific SARS-CoV-2 Antigens in Human Specimens Using Direct/Enrichment Dual-Mode Fluorescence Lateral Flow Immunoassay

Abstract
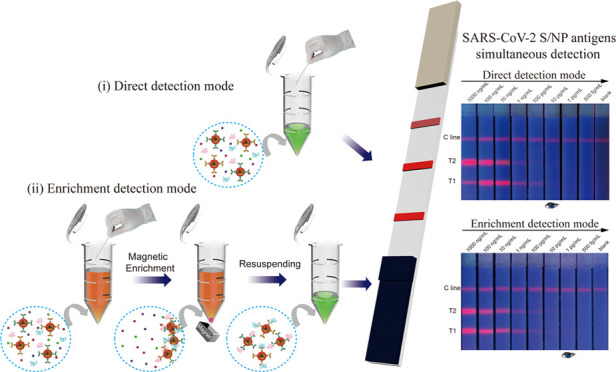
Sensitive point-of-care methods for detecting severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) antigens in clinical specimens are urgently needed to achieve rapid screening of viral infection. We developed a magnetic quantum dot-based dual-mode lateral flow immunoassay (LFIA) biosensor for the high-sensitivity simultaneous detection of SARS-CoV-2 spike (S) and nucleocapsid protein (NP) antigens, which is beneficial for improving the detection accuracy and efficiency of SARS-CoV-2 infection in the point-of-care testing area. A high-performance magnetic quantum dot with a triple-QD shell (MagTQD) nanotag was first fabricated and integrated into the LFIA system to provide superior fluorescence signals, enrichment ability, and detectability for S/NP antigen testing. Two detection modes were provided by the proposed MagTQD-LFIA. The direct mode was used for rapid screening or urgent detection of suspected samples within 10 min, and the enrichment mode was used for the highly sensitive and quantitative analysis of SARS-CoV-2 antigens in biological samples without the interference of the “hook effect.” The simultaneous detection of SARS-CoV-2 S/NP antigens was conducted in one LFIA strip, and the detection limits for two antigens under direct and enrichment modes were 1 and 0.5 pg/mL, respectively. The MagTQD-LFIA showed high accuracy, specificity, and stability in saliva and nasal swab samples and is an efficient tool with flexibility to meet the testing requirements for SARS-CoV-2 antigens in various situations.
Related collections
Most cited references52

- Record: found
- Abstract: found
- Article: found
The species Severe acute respiratory syndrome-related coronavirus : classifying 2019-nCoV and naming it SARS-CoV-2
- Record: found
- Abstract: found
- Article: not found
Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor
- Record: found
- Abstract: found
- Article: not found