- Record: found
- Abstract: found
- Article: found
Viral Characteristics Associated with the Clinical Nonprogressor Phenotype Are Inherited by Viruses from a Cluster of HIV-1 Elite Controllers
Read this article at
ABSTRACT
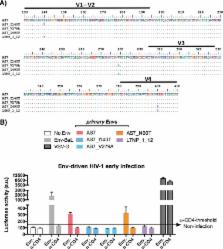
A small group of HIV-1-infected individuals, called long-term nonprogressors (LTNPs), and in particular a subgroup of LTNPs, elite controllers (LTNP-ECs), display permanent control of viral replication and lack of clinical progression. This control is the result of a complex interaction of host, immune, and viral factors. We identified, by phylogenetic analysis, a cluster of LTNP-ECs infected with very similar low-replication HIV-1 viruses, suggesting the contribution of common viral features to the clinical LTNP-EC phenotype. HIV-1 envelope (Env) glycoprotein mediates signaling and promotes HIV-1 fusion, entry, and infection, being a key factor of viral fitness in vitro, cytopathicity, and infection progression in vivo. Therefore, we isolated full-length env genes from viruses of these patients and from chronically infected control individuals. Functional characterization of the initial events of the viral infection showed that Envs from the LTNP-ECs were ineffective in the binding to CD4 and in the key triggering of actin/tubulin-cytoskeleton modifications compared to Envs from chronic patients. The viral properties of the cluster viruses result in a defective viral fusion, entry, and infection, and these properties were inherited by every virus of the cluster. Therefore, inefficient HIV-1 Env functions and signaling defects may contribute to the low viral replication capacity and transmissibility of the cluster viruses, suggesting a direct role in the LTNP-EC phenotype of these individuals. These results highlight the important role of viral characteristics in the LTNP-EC clinical phenotype. These Env viral properties were common to all the cluster viruses and thus support the heritability of the viral characteristics.
IMPORTANCE
HIV-1 long-term nonprogressor elite controller patients, due to their permanent control of viral replication, have been the object of numerous studies to identify the factors responsible for this clinical phenotype. In this work, we analyzed the viral characteristics of the envelopes of viruses from a phylogenetic cluster of LTNP-EC patients. These envelopes showed ineffective binding to CD4 and the subsequent signaling activity to modify actin/tubulin cytoskeletons, which result in low fusion and deficient entry and infection capacities. These Env viral characteristics could explain the nonprogressor clinical phenotype of these patients. In addition, these inefficient env viral properties were present in all viruses of the cluster, supporting the heritability of the viral phenotype.
Related collections
Most cited references39
- Record: found
- Abstract: found
- Article: not found
HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors.
- Record: found
- Abstract: found
- Article: not found
HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy.
- Record: found
- Abstract: found
- Article: not found