- Record: found
- Abstract: found
- Article: found
Transposon-induced methylation of the RsMYB1 promoter disturbs anthocyanin accumulation in red-fleshed radish
Read this article at
Abstract
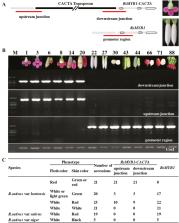
A CACTA transposon-induced methylation of the RsMYB1 promoter might be associated with the production of white-fleshed mutants in red-fleshed radish.
Abstract
Red-fleshed radish ( Raphanus sativus L.) is a unique cultivar whose taproot is rich in anthocyanins beneficial to human health. However, the frequent occurrence of white-fleshed mutants affects the purity of commercially produced radish and the underlying mechanism has puzzled breeders for many years. In this study, we combined quantitative trait location by genome resequencing and transcriptome analyses to identify a candidate gene ( RsMYB1) responsible for anthocyanin accumulation in red-fleshed radish. However, no sequence variation was found in the coding and regulatory regions of the RsMYB1 genes of red-fleshed (MTH01) and white-fleshed (JC01) lines, and a 7372 bp CACTA transposon in the RsMYB1 promoter region occurred in both lines. A subsequent analysis suggested that the white-fleshed mutant was the result of altered DNA methylation in the RsMYB1 promoter. This heritable epigenetic change was due to the hypermethylated CACTA transposon, which induced the spreading of DNA methylation to the promoter region of RsMYB1. Thus, RsMYB1 expression was considerably down-regulated, which inhibited anthocyanin biosynthesis in the white-fleshed mutant. An examination of transgenic radish calli and the results of a virus-induced gene silencing experiment confirmed that RsMYB1 is responsible for anthocyanin accumulation. Moreover, the mutant phenotype was partially eliminated by treatment with a demethylating agent. This study explains the molecular mechanism regulating the appearance of white-fleshed mutants of red-fleshed radish.
Related collections
Most cited references48
- Record: found
- Abstract: found
- Article: not found
Rapid isolation of high molecular weight plant DNA.
- Record: found
- Abstract: found
- Article: not found
QTL-seq: rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations.
- Record: found
- Abstract: found
- Article: not found