- Record: found
- Abstract: found
- Article: found
Rat post-implantation epiblast-derived pluripotent stem cells produce functional germ cells

Read this article at
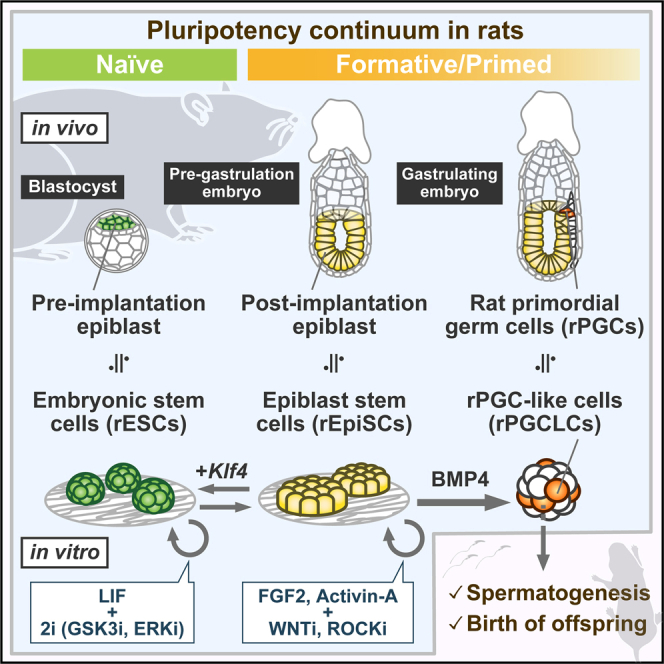
Summary
In mammals, pluripotent cells transit through a continuum of distinct molecular and functional states en route to initiating lineage specification. Capturing pluripotent stem cells (PSCs) mirroring in vivo pluripotent states provides accessible in vitro models to study the pluripotency program and mechanisms underlying lineage restriction. Here, we develop optimal culture conditions to derive and propagate post-implantation epiblast-derived PSCs (EpiSCs) in rats, a valuable model for biomedical research. We show that rat EpiSCs (rEpiSCs) can be reset toward the naive pluripotent state with exogenous Klf4, albeit not with the other five candidate genes ( Nanog, Klf2, Esrrb, Tfcp2l1, and Tbx3) effective in mice. Finally, we demonstrate that rat EpiSCs retain competency to produce authentic primordial germ cell-like cells that undergo functional gametogenesis leading to the birth of viable offspring. Our findings in the rat model uncover principles underpinning pluripotency and germline competency across species.
Graphical abstract
Highlights
Motivation
Pluripotent stem cells (PSCs) that mirror the pluripotent state in the pre-gastrulation epiblast allow us to examine the molecular mechanisms underlying pluripotent transitions and the differentiation of germ line and soma. Despite rats being a prominent animal model for biomedical research, alongside mice, there has been limited investigation into the derivation and characterization of rat PSCs derived from the post-implantation epiblast (rEpiSCs). This study explores the optimal culture conditions for successfully deriving and expanding rEpiSCs. Additionally, we investigate the molecular characteristics of established rEpiSCs, their ability to reset to a naive pluripotent state, and their competence in producing functional germ cells.
Abstract
Iwatsuki et al. develop optimal culture conditions for rat epiblast-derived pluripotent stem cells (EpiSCs). These rEpiSCs exhibit molecular similarities to pre-gastrulating pluripotent epiblasts and demonstrate the ability to generate functional germ cells. Therefore, the approach provides valuable tools for studying pluripotency and in vitro gametogenesis.
Related collections
Most cited references56
- Record: found
- Abstract: found
- Article: not found
STAR: ultrafast universal RNA-seq aligner.
- Record: found
- Abstract: found
- Article: not found
Comprehensive Integration of Single-Cell Data
- Record: found
- Abstract: found
- Article: not found