- Record: found
- Abstract: found
- Article: found
Clinical application of chromosomal microarray analysis in fetuses with increased nuchal translucency and normal karyotype
Read this article at
Abstract
Background
Submicroscopic chromosomal imbalance is associated with an increased nuchal translucency (NT). Most previous research has recommended the use of chromosomal microarray analysis (CMA) for prenatal diagnosis if the NT ≥ 3.5 mm. However, there is no current global consensus on the cutoff value for CMA. In this study, we aimed to discuss the fetuses with smaller increased NT which was between cutoff value of NT for karyotype analysis (NT of 2.5 mm in China) and the recommended cutoff value for CMA (NT of 3.5 mm) whether should be excluded from CMA test.
Methods
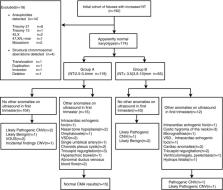
Singleton pregnant women ( N = 192) who had undergone invasive procedures owing to an increased NT (NT ≥ 2.5 mm) were enrolled. Fetal cells were collected and subjected to single nucleotide polymorphism array and karyotype analyses simultaneously. Cases were excluded if the karyotype analysis indicated aneuploidy and apparent structural aberrations.
Results
Fourteen cases of aneuploidy and four cases of structural abnormalities were excluded. Of the remaining 174 cases, 119 fetuses had NTs of 2.5–3.4 mm, and 55 fetuses with NT ≥ 3.5 mm. Eleven copy number variants (CNVs) were identified. In fetuses with smaller NTs, six (6/119, 5.9%) variations were detected, including two (2/119, 1.6%) clinically significant CNVs (pathogenic or likely pathogenic CNV), one likely benign CNV, two variants unknown significance, and one incidental CNV. Five (5/55, 9.1%) variations were found in fetuses with NT ≥ 3.5 mm. Among these CNVs, three (3/55, 5.5%) cases had clinically significant CNVs, and two had likely benign CNV. There were no statistically significant differences in the incidence of all CNVs and clinically significant CNVs in the two groups ( p > 0.05).
Related collections
Most cited references22
- Record: found
- Abstract: found
- Article: not found
Mechanisms of mosaicism, chimerism and uniparental disomy identified by single nucleotide polymorphism array analysis.
- Record: found
- Abstract: found
- Article: not found
