- Record: found
- Abstract: found
- Article: found
New Insectotoxin from Tibellus Oblongus Spider Venom Presents Novel Adaptation of ICK Fold
Read this article at
Abstract
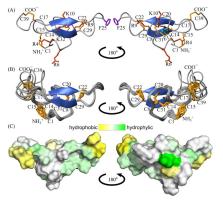
The Tibellus oblongus spider is an active predator that does not spin webs and remains poorly investigated in terms of venom composition. Here, we present a new toxin, named Tbo-IT2, predicted by cDNA analysis of venom glands transcriptome. The presence of Tbo-IT2 in the venom was confirmed by proteomic analyses using the LC-MS and MS/MS techniques. The distinctive features of Tbo-IT2 are the low similarity of primary structure with known animal toxins and the unusual motif of 10 cysteine residues distribution. Recombinant Tbo-IT2 (rTbo-IT2), produced in E. coli using the thioredoxin fusion protein strategy, was structurally and functionally studied. rTbo-IT2 showed insecticidal activity on larvae of the housefly Musca domestica (LD 100 200 μg/g) and no activity on the panel of expressed neuronal receptors and ion channels. The spatial structure of the peptide was determined in a water solution by NMR spectroscopy. The Tbo-IT2 structure is a new example of evolutionary adaptation of a well-known inhibitor cystine knot (ICK) fold to 5 disulfide bonds configuration, which determines additional conformational stability and gives opportunities for insectotoxicity and probably some other interesting features.
Related collections
Most cited references69
- Record: found
- Abstract: found
- Article: not found
NMRPipe: a multidimensional spectral processing system based on UNIX pipes.
- Record: found
- Abstract: found
- Article: not found
A Cross-platform Toolkit for Mass Spectrometry and Proteomics
- Record: found
- Abstract: found
- Article: not found