- Record: found
- Abstract: found
- Article: found
Information integration during bioelectric regulation of morphogenesis of the embryonic frog brain

Read this article at
Summary
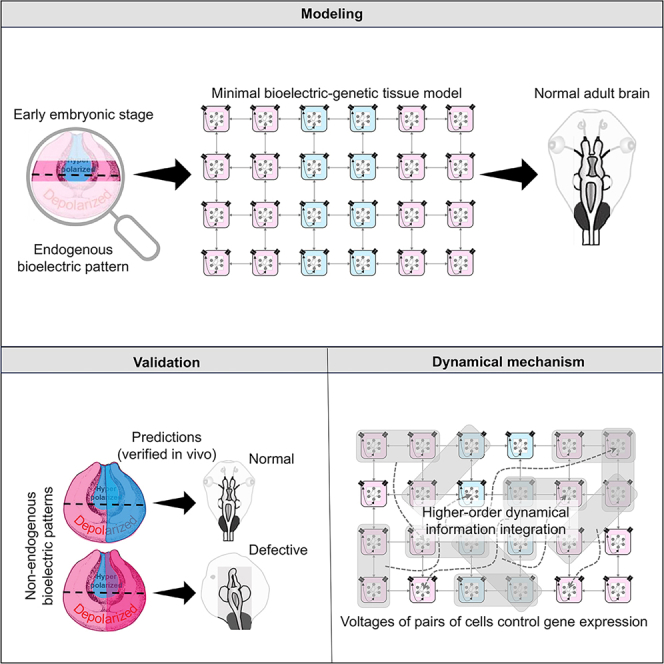
Spatiotemporal patterns of cellular resting potential regulate several aspects of development. One key aspect of the bioelectric code is that transcriptional and morphogenetic states are determined not by local, single-cell, voltage levels but by specific distributions of voltage across cell sheets. We constructed and analyzed a minimal dynamical model of collective gene expression in cells based on inputs of multicellular voltage patterns. Causal integration analysis revealed a higher-order mechanism by which information about the voltage pattern was spatiotemporally integrated into gene activity, as well as a division of labor among and between the bioelectric and genetic components. We tested and confirmed predictions of this model in a system in which bioelectric control of morphogenesis regulates gene expression and organogenesis: the embryonic brain of the frog Xenopus laevis. This study demonstrates that machine learning and computational integration approaches can advance our understanding of the information-processing underlying morphogenetic decision-making, with a potential for other applications in developmental biology and regenerative medicine.
Graphical abstract
Highlights
-
•
A minimal model shows how cells can sense large-scale patterns of cell voltage
-
•
Model makes predictions about the outcome of new bioelectric patterns
-
•
Model predictions are verified by experiments in Xenopus brain development
-
•
Higher-order information integration is seen in voltage-transcription dynamics
Abstract
Biomimetics; Embryology; Complex system biology; In silico biology; Model organism
Related collections
Most cited references194
- Record: found
- Abstract: found
- Article: not found
Fiji: an open-source platform for biological-image analysis.
- Record: found
- Abstract: not found
- Article: not found
Physics-Informed Neural Networks: A Deep Learning Framework for Solving Forward and Inverse Problems Involving Nonlinear Partial Differential Equations
- Record: found
- Abstract: found
- Article: not found