- Record: found
- Abstract: found
- Article: found
AutoSOME: a clustering method for identifying gene expression modules without prior knowledge of cluster number
Read this article at
Abstract
Background
Clustering the information content of large high-dimensional gene expression datasets has widespread application in "omics" biology. Unfortunately, the underlying structure of these natural datasets is often fuzzy, and the computational identification of data clusters generally requires knowledge about cluster number and geometry.
Results
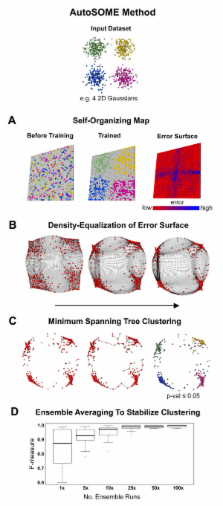
We integrated strategies from machine learning, cartography, and graph theory into a new informatics method for automatically clustering self-organizing map ensembles of high-dimensional data. Our new method, called AutoSOME, readily identifies discrete and fuzzy data clusters without prior knowledge of cluster number or structure in diverse datasets including whole genome microarray data. Visualization of AutoSOME output using network diagrams and differential heat maps reveals unexpected variation among well-characterized cancer cell lines. Co-expression analysis of data from human embryonic and induced pluripotent stem cells using AutoSOME identifies >3400 up-regulated genes associated with pluripotency, and indicates that a recently identified protein-protein interaction network characterizing pluripotency was underestimated by a factor of four.
Conclusions
By effectively extracting important information from high-dimensional microarray data without prior knowledge or the need for data filtration, AutoSOME can yield systems-level insights from whole genome microarray expression studies. Due to its generality, this new method should also have practical utility for a variety of data-intensive applications, including the results of deep sequencing experiments. AutoSOME is available for download at http://jimcooperlab.mcdb.ucsb.edu/autosome.
Related collections
Most cited references32
- Record: found
- Abstract: found
- Article: not found
DAVID: Database for Annotation, Visualization, and Integrated Discovery.
- Record: found
- Abstract: not found
- Article: not found
Molecular Classification of Cancer: Class Discovery and Class Prediction by Gene Expression Monitoring
- Record: found
- Abstract: found
- Article: not found