- Record: found
- Abstract: found
- Article: found
Long noncoding RNA ZEB1-AS1 epigenetically regulates the expressions of ZEB1 and downstream molecules in prostate cancer
Read this article at
Abstract
Background
Emerging studies show that long noncoding RNAs (lncRNAs) play important roles in carcinogenesis and cancer progression. The lncRNA ZEB1 antisense 1 (ZEB1-AS1) derives from the promoter region of ZEB1 and we still know little about its expressions, roles and mechanisms.
Methods
RACE was used to obtain the sequence of ZEB1-AS1. RNA interference was used to decrease ZEB1-AS1 expression. Adenovirus expression vector was used to increase ZEB1-AS1 expression. CHIP and RIP were used to detect the epigenetic mechanisms by which ZEB1-AS1 regulated ZEB1. CCK8 assay, wound healing assay and transwell assay were used to measure proliferation and migration of prostate cancer cells.
Results
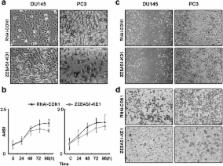
In this study, in prostate cancer cells, we found that RNAi-mediated downregulation of ZEB1-AS1 induced significant ZEB1 inhibition while artificial overexpression of ZEB1-AS1 rescued ZEB1 expression, which means that ZEB1-AS1 promotes ZEB1 expression. Also, ZEB1-AS1 indirectly inhibited miR200c, the well-known target of ZEB1, and upregulated miR200c’s target BMI1. Mechanistically, ZEB1-AS1 bound and recruited histone methyltransferase MLL1 to the promoter region of ZEB1, induced H3K4me3 modification therein, and activated ZEB1 transcription. Biologically, ZEB1-AS1 promoted proliferation and migration of prostate cancer cells.
Related collections
Most cited references28
- Record: found
- Abstract: found
- Article: not found
Induced ncRNAs Allosterically Modify RNA Binding Proteins in cis to Inhibit Transcription
- Record: found
- Abstract: found
- Article: not found
Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4.
- Record: found
- Abstract: found
- Article: not found