- Record: found
- Abstract: found
- Article: found
A toolkit of tagged glp-1 alleles reveals strong glp-1 expression in the germline, embryo, and spermatheca
brief-report
Erika B Sorensen
1
,
§ ,
Hannah S Seidel
2
,
§ ,
Sarah L Crittenden
3 ,
Joseph H Ballard
1 ,
Judith Kimble
3
,
§
Erin Cram (Reviewer)
22 June 2020
2020
Read this article at
There is no author summary for this article yet. Authors can add summaries to their articles on ScienceOpen to make them more accessible to a non-specialist audience.
Abstract
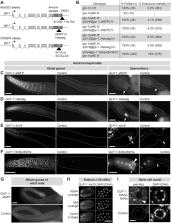
Figure 1
Tagged glp-1 alleles are expressed in the distal germline, embryo, and spermatheca.
(A) Schematic of tagged glp-1 alleles. (B) Rescue of glp-1 null phenotype by MosSCI
glp-1 alleles. (C-F) Adult hermaphrodite gonads, oriented distal to the left. Arrow,
spermatheca. (F) Arrowhead, GLP-1::6xMyc6xHis visible within spermathecal cell nucleus.
Staining in oocyte nuclei likely includes a large component of background staining
because staining is present at similar levels in controls. (G) Adult male gonads.
(C-G) Dashed line, outline of gonad. (H) Embryo expressing GLP-1::4xV5. Images are
maximum-intensity z-projections through equal z-ranges of the same embryo. Anti-V5
staining in control embryos (not shown) was uniformly dim. (C-H) Scale bar, 20 µm.
(I) Distal-most germ cell nuclei of adult hermaphrodite. Dashed line, germ cell nucleus.
Arrowhead, GLP-1::6xMyc6xHis visible within germ cell nucleus. Scale bar, 2 µm. (C-G,
I) Control, animals lacking a tagged glp-1 allele.
Description
The C. elegans genome encodes two Notch receptors, GLP-1 and LIN-12 (Greenwald and
Kovall, 2013). These receptors function in multiple tissues to regulate development,
behavior, and reproduction. GLP-1 controls cell fate decisions in the germline (Kimble
and Seidel, 2013) and early embryo (Priess, 2005). LIN-12 controls development of
the vulva (Greenwald, 2005; Sternberg, 2005). These receptors also act outside the
context of development in post-mitotic neurons to regulate chemosensation (Singh et
al., 2011), locomotion (Chao et al., 2005), and life-history decisions (Ouellet et
al., 2008). Additional processes regulated by Notch signaling include oocyte growth
(Nadarajan et al., 2009) and ovulation (McGovern et al., 2018). Understanding the
many functions of Notch signaling therefore requires a global picture of Notch receptor
expression.
Previous efforts to visualize Notch receptor expression have focused on lin-12 expression
in the soma and glp-1 expression in the germline and early embryo. LIN-12 has been
visualized successfully using LIN-12::GFP fusion proteins introduced via traditional,
multi-copy transgenic techniques (e.g. Levitan and Greenwald, 1998; Sarov et al.,
2012). Such techniques have been difficult for visualizing glp-1 expression in the
germline, until recently (Cinquin et al., 2015; Gutnik et al., 2018), due to germline
silencing of multi-copy transgenes (Kelly et al., 1997; Merritt and Seydoux, 2010).
Visualizing GLP-1expression in the germline has therefore relied on antibody staining,
primarily using an antibody raised against the GLP-1 extra-cellular domain (Crittenden
et al., 1994). This antibody reveals easily detectable GLP-1 protein at the plasma
membrane in the distal germline and early embryos (Crittenden et al., 1994; Evans
et al., 1994), but it does not allow visualization of the GLP-1 nuclear intracellular
domain (NICD), which moves into the nucleus upon receptor activation. This antibody
is therefore not useful for identifying cells that have directly received active GLP-1
signaling. To overcome this limitation, and to broaden the toolkit for visualizing
glp-1 expression, we created five strains expressing tagged glp-1 alleles, including
transgenes and CRISPR tags at the endogenous locus. We report here the expression
patterns of the tagged glp-1 alleles, focusing on the adult gonad.
We created five tagged glp-1 alleles, each expressing GLP-1 protein tagged with one
of the following tags: sfGFP (~27 kDa), Halotag (~33 kDa), 4xV5 (~7 kDa), 3xOLLAS
(~4.4 kDa), or 6xMyc6xHis (~8 kDa). The 6xMyc6xHis tag was placed at the C-terminus
of glp-1, downstream of the GLP-1 PEST domain (Figure 1A). The remaining tags were
placed C-terminal to the GLP-1 Ankyrin repeats but upstream of the PEST domain (Figure
1A). These sites place the tags at or near the C-terminus of the NICD. Strains expressing
glp-1::sfGFP, glp-1::Halotag, and glp-1::6xMyc6xHis were created via Mos1-mediated
single-copy insertion of the transgene into the genome. Strains expressing glp-1::4xV5
and glp-1::3xOLLAS were created by CRISPR-mediated insertion of the tag into the endogenous
glp-1 locus.
To assess functionality of the tagged GLP-1 proteins, we tested each for rescue of
the glp-1 loss-of-function phenotype, which includes infertility and maternal-effect
embryonic lethality. Rescue by glp-1::sfGFP, glp-1::Halotag, and glp-1::6xMyc6xHis
was assessed by crossing each transgene into animals carrying the null allele glp-1(q46)
(Kodoyianni et al., 1992). Rescue by the CRISPR-generated alleles (glp-1::4xV5 and
glp-1::3xOLLAS)was assessed by examining animals homozygous for the tagged allele.
We observed strong rescue for all tagged glp-1 alleles: Animals expressing glp-1::4xV5
or glp-1::3xOLLAS were all fertile (n > 50 hermaphrodites per allele) and showed no
noticeable embryonic lethality; animals expressing glp-1::6xMyc6xHis,
glp-1::sfGFP or glp-1::Halotag were all fertile, but showed a low penetrance embryonic
lethality (Figure 1B). We conclude that all five tagged glp-1 alleles encode functional
GLP-1 proteins.
We characterized expression of the tagged glp-1 alleles in adult hermaphrodites and
males, focusing on the gonad. GLP-1::6xMyc6xHis, GLP-1::4xV5, and GLP-1::3xOLLAS were
visualized with immunostaining of extruded gonads. GLP-1::sfGFP and GLP-1::Halotagwere
examined in live animals and extruded gonads. All five tagged alleles showed expression
in two areas: the distal germline and spermatheca (Figure 1C-G). Signal in the spermatheca
was strongest at the membrane but also sometimes visible at a lower level in spermathecal
nuclei (Figure 1F). Expression in the distal germline was present in both sexes and
strongest in the distal-most ~20 rows of germ cells, becoming weaker more proximally
(Figure 1C-G). Germ cells in the distal-most gonad showed membrane staining similar
to that seen with antibodies to the extracellular domain of GLP-1 (e.g. Crittenden
et al., 1994); in addition, staining in the nucleus was detected, but at a low level
compared to membrane staining (Figure 1I). This difference in membrane and intranuclear
GLP-1 staining is consistent with a previous report of GLP-1 NICD expression in the
distal germline (Gutnik et al., 2018). We also examined pre-gastrulation embryos and
observed both membrane and nuclear expression of tagged GLP-1, in a pattern consistent
with GLP-1 expression in the AB cell lineage (Figure 1H), as reported previously (Evans
et al., 1994; Priess, 2005). Expression in L4 and adult animals was not observed outside
the gonad or in the male somatic gonad (although we cannot exclude the possibility
of expression below our threshold of detection). Spatial patterns of expression were
essentially the same for all five tagged glp-1 alleles, although signal-to-background
ratios differed: 4xV5 and Halotag gave strong signal in animals with the tagged glp-1
alleles and minimal background in controls; sfGFP gave a dimmer signal than 4xV5 and
Halotag; and 3xOLLAS and 6xMyc6xHis had higher background in controls, including in
germ cell nuclei (data not shown for 3xOLLAS). These results show that the sites of
strongest glp-1 expression in adults are the distal germline and the spermatheca.
Our study provides a toolkit of five tagged glp-1 alleles. These alleles confirm the
expected pattern of GLP-1 expression in the distal germline (Crittenden et al., 1994),
and they further enable visualization of the GLP-1 NICD in germ cell nuclei (Figure
1I). These alleles also show strong expression in an unexpected site: the spermatheca
(Figure 1C-F). GLP-1 has not been reported previously in the spermatheca. Yet Notch
signaling has been proposed to affect passage of the oocyte through the spermatheca
because mutations in the Notch ligand apx-1 cause defects in ovulation (McGovern et
al., 2018). In addition, the DNA-binding co-factor of Notch, LAG-1, is strongly expressed
in spermathecal nuclei (Chen et al., 2020). Notch signaling in the spermatheca was
proposed to be mediated by LIN-12 (McGovern et al., 2018), but our study suggests
that GLP-1 is another reasonable candidate. We expect that our toolkit of tagged glp-1
alleles will prove useful in visualizing GLP-1 in the germ cells, as well as in investigating
a possible role for GLP-1 in the spermatheca.
Methods
Methods
Strains and growth conditions
Worms were grown at 20°C on standard nematode growth media plates seeded with E. coli
OP50.
N2
EG8081 unc-119(ed3) III; oxTi177[ttTi5605 + NeoR(+) + unc-18(+) ] IV
EG6699 ttTi5605 II; unc-119(ed3) III; oxEx1578
JK4862 glp-1(q46) III / hT2[bli-4(e937) let-?(q782) qIs48] (I;III)
JK5008 qSi44[Cbr-unc-119 + glp-1::6xMyc6xHis] II ; glp-1(q46) III
JK5525 glp-1(q46) III; qSi155[Cbr-unc-119 + glp-1::Halotag] IV
JK5526 glp-1(q46) III; qSi156[Cbr-unc-119 + glp-1::Halotag] IV
JK5535 glp-1(q46) III; qSi246[Cbr-unc-119 + glp-1::sfGFP] IV
JK5548 glp-1(q46) III; qSi244[Cbr-unc-119 + glp-1::sfGFP] IV
JK5973 glp-1(q997[glp-1::3xOLLAS]) III
JK5933 glp-1(q1000[glp-1::4xV5]) III
Creation of glp-1::sfGFP, glp-1::Halotag, and glp-1::6xMyc6xHis constructs
glp-1::sfGFP and glp-1::Halotag were constructed by cloning the genomic glp-1 locus
into MosSCI vector pCFJ151 (Frøkjaer-Jensen et al., 2008), using Gibson assembly (Gibson,
2011). This construct contains 2,506 bp upstream of the glp-1 start codon and 934
bp downstream of the glp-1 stop codon. sfGFP (Pédelacq et al., 2006) and Halotag (Los
et al., 2008) were amplified from sfGFP expression plasmid (Sandia Biotech #23004006)
and pFN18A (Promega #G2751), respectively, using primers containing NruI sites in
primer tails. sfGFP and Halotag were inserted into glp-1 at the NruI site using standard
ligation. glp-1::6xMyc6xHis was constructed by cloning the same genomic glp-1 locus
into MosSCI vector pCFJ151, using Gibson assembly, except that unique NotI and PmeI
sites were inserted before the stop codon using primers containing NotI and PmeI sites
in primer tails. The 6xMyc6xHis tag was amplified from plasmid 4myc-deltaE pAdtet7
(Sorensen and Conner, 2010) and inserted between the NotI and PmeI sites using standard
ligation.
Single-copy insertion of glp-1::sfGFP, glp-1::Halotag, and glp-1::6xMyc6xHis
glp-1::sfGFP and glp-1::Halotag were integrated into site oxTi177 on LGIV by injection
of their respective constructs into strain EG8081 using the Universal MosSCI technique,
as described (Frøkjær-Jensen et al., 2014). glp-1::6xMyc6xHis was integrated into
site ttTi5605 site on LGII by injection of the glp-1::6xMyc6xHis construct into strain
EG6699, as described (Frøkjær-Jensen et al., 2012). Injection mix contained 15 ng/µl
glp-1::sfGFP or glp-1::Halotag or glp-1::6xMyc6xHis, 50 ng/µl pCFJ601, 10 ng/µl pGH8,
25 ng/µl pCFJ90, 5 ng/µl pCFJ104, and sometimes 10 ng/µl pMA122. Positive insertions
were recovered based on the Non-Unc phenotype and loss of co-injection markers. Insertions
were crossed into a glp-1(q46) genetic background. Homozygosity of the glp-1(q46)
allele was confirmed using primers that amplify only the endogenous locus of glp-1
(outer forward, 5’-aaacactttttgggtgctgtg-3’; inner forward, 5’-gatttgaactgccatgatttat-3’;
inner reverse, 5’-acagcttgccgatacctgc-3’; outer reverse, 5’-tcagttcattgatcttgtcgacac-3’)
followed by digestion with MseI.
CRISPR/Cas9 genome editing to create glp-1::4xV5 and glp-1::3xOLLAS
glp-1::4xV5 and glp-1::3xOLLAS were generated via a co-CRISPR genome editing strategy
using a CRISPR/Cas9 RNA-protein complex (Arribere et al., 2014; Paix et al., 2015).
Wildtype animals were injected with a mix containing a gene-specific crRNA (10 μM,
IDT-Alt-RTM), unc-58 co-CRISPR crRNA (4 μM, IDT-Alt-RTM), tracrRNA (13.6 μM, IDT-Alt-RTM),
gene specific repair oligo (4 μM), unc-58 repair oligo (1.34 μM), and Cas-9 protein
(24.5 μM). F1 progeny of injected hermaphrodites were screened for desired mutations
by PCR and Sanger sequencing. Each allele was outcrossed against wild-type twice prior
to analysis. The glp-1-specific crRNA was 5’-CAG GUC AUG GUG CUA AGU CUG UUU UAG AGC
UAU GCU-3’. The repair oligo for glp-1::4xV5 was 5’-GCT CTT TTG ATA TTC TTC ACA GTT
TGT CGC CCA GAG GTG CTA TCA AGT CCG AGG AGT GGG TTA GGG ATA GGC TTA CCG GTG CTA TCA
AGT CCG AGG AGT GGG TTT GGG ATT GGC TTT CCA GTA CTA TCT AGA CCG AGG AGA GGG TTA GGG
ATA GGC TTA CCC TTA GCA CCA TGA CCT GAC TTG ACT ATT TGC TG-3’. The repair oligo for
glp-1::4xV5 contained three copies of the V5 tag, but the insertion recovered contained
four copies of the V5 tag. The repair oligo for glp-1::3xOLLAS was 3’-GCT CTT TTG
ATA TTC TTC ACA GTT TGT CGT CCA GAC TTT CCC ATG AGA CGT GGT CCG AGC TCG TTG GCG AAT
CCG GAT TGC TTT CCC ATG AGA CGT GGT CCG AGC TCG TTG GCG AAT CCG GAT TGC TTT CCC ATG
AGA CGT GGT CCG AGC TCG TTG GCG AAT CCG GAC TTA GCA CCA TGA CCT GAC TTG ACT ATT TGC
TG-3’. Primers used to genotype animals for insertions were SLR10 (5’-AACTCTGTGGGGACCAGTG-3’)
and SLR21 (5’-GATGCTAGCTGTTCGTGC-3’).
Locations of tags within the GLP-1 protein sequence
The 4xV5 and 3xOLLAS tags were inserted between lysine 1185 and serine 1186 (…SGHGAK-tag-SGRQTV…).
sfGFP and Halotag were inserted between serine 1209 and arginine 1210 (…TSAASS-tag-RETNHL…).
The 6xMyc6xHis tag was inserted immediately upstream of the stop codon (…NGSFYC-tag-*).
Rescue of glp-1(q46)
Fertility was assessed by examining hermaphrodites aged 24-hrs post mid-L4 at 400X
magnification. Animals were scored as ‘fertile’ if embryos were present in both sides
of the uterus and if both arms of the gonad were similar in size and morphology to
those of wildtype animals. Embryo viability was assessed by allowing hermaphrodites
aged 24-hrs post mid-L4 to lay eggs for ~8 hrs. Embryos were scored as ‘viable’ if
they hatched within ~16 hours after removal of the parent animal from the plate.
Fixation, staining, and imaging
Animals were collected as L4s or as adults aged 24 hours post mid-L4. In all experiments,
animals expressing tagged glp-1 alleles were treated in parallel with control animals
lacking the allele.
For glp-1::4xV5 and glp-1::3xOLLAS, gonads were extruded in 0.25 mM levamisole/PBSTw
(PBS + 0.1% Tween-20) and fixed in 4% (w/v) paraformaldehyde/PBSTw for 10 min at room
temperature. Gonads were permeabilized in 0.1%Triton X-100/PBSTw for 30 min and blocked
in PBSB (PBSTw + 0.5% BSA) for 20 min at room temperature. Gonads were stained overnight
at 4°C in mouse anti-V5 (Bio-Rad #MCA1360) or rat anti-OLLAS(Novus #NBP1-06713) diluted
1:1000 in block. Gonads were washed three times in block for 10 min per wash and incubated
for 1 hour at room temperature in 0.1 µg/ml DAPI and AlexaFluor 555 donkey anti-mouse
(Invitrogen #A31570), AlexaFluor 647 donkey anti-mouse (Invitrogen #A31571), or AlexaFluor
488 donkey anti-rat (Invitrogen #A21208) diluted 1:1000 in block. Gonads were washed
as before and mounted in ProLong Gold (Thermo-Fisher #P36934). For staining of embryos,
gravid hermaphrodites were cut on a subbed slide in PBS to release embryos. Paraformaldehyde
was added to 4% (w/v), and embryos were depressed under a coverslip to crack the eggshell.
The slide was incubated in a moist chamber for 30 min at room temperature and frozen
on dry ice for 5-10 min. The coverslip was removed, and the slide was immersed in
-20ºC methanol for 10 min, followed by room temperature methanol for 10 min. Embryos
were blocked and stained with antibodies as described for gonads.
For glp-1::6xMyc6xHis, gonads were extruded in 0.2 mM levamisole/PBSTw and fixed in
3% (w/v) paraformaldehyde/0.1 M K2HPO4 (pH 7.2) for 20 min at room temperature. Gonads
were washed once in PBSTw and permeabilized in -20°C methanol for 5 min. Gonads were
washed three times in PBSTw and blocked in PBSB for 45 min at room temperature. Gonads
were stained overnight at 4°C in mouse monoclonal antibody 9E10 diluted 1:10 in block.
The 9E10 antibody, developed by Michael J. Bishop, was obtained from the Developmental
Studies Hybridoma Bank, created by the NICHD of the NIH (The University of Iowa, Department
of Biology, Iowa City, IA 52242). Gonads were washed four times in PBSTw for 15 min
total and incubated for 1 hour at room temperature in 0.1 µg/ml DAPI and AlexaFluor
555 goat anti-mouse (Thermo-Fisher #A28180) diluted 1:1000 in block. Gonads were washed
as before and mounded in Vectashield (Vectorlabs #H-1000).
For glp-1::sfGFP, whole living animals were imaged on agarose pads. For imaging of
gonads, gonads were extruded, fixed, and permeabilized as for glp-1::6xMyc6xHis. Gonads
were washed three times in PBSTw and mounted in Vectashield.
For glp-1::Halotag, animals were incubated for 90 min in HaloTag® TMR Ligand (Promega,
#G8252) diluted 1:1000 in M9/0.1% Tween-20. Animals were rinsed once with M9/0.1%
Tween-20 and allowed to crawl around for 30 min on a seeded plate in the dark. For
imaging of whole animals, live animals were mounted on agarose pads and imaged immediately.
For imaging of gonads, gonads were extruded in 0.2 mM levamisole/PBSTw and fixed in
3% (w/v) paraformaldehyde/0.1 M K2HPO4 (pH 7.2) for 20 min at room temperature. Gonads
were mounted in Vectashield. For glp-1::Halotag embryos (images not shown), gravid
hermaphrodites were cut on a subbed slide in PBS/100 nM HaloTag® TMR Ligand to release
embryos. Paraformaldehyde was added to 4% (w/v), and embryos were incubated for 10
min in a moist chamber. Embryos were depressed under a coverslip to crack the eggshell,
and the slide was frozen on dry ice for 5-10 min. The coverslip was removed, and the
slide was washed in PBSTw. Embryos were mounted in ProLong Gold.
Images of glp-1::sfGFP, glp-1::Halotag, and glp-1::6xMyc6xHis (spermatheca) were acquired
on a Zeiss AxioScope A1 microscope equipped with a AxioCam 503 mono camera. Images
of glp-1::4xV5 and glp-1::6xMyc6xHis (distal germline) were acquired on a Leica SP8
laser scanning confocal microscope. For each set of images, brightness levels were
adjusted the same for experimental and control images.
Related collections
Most cited references21
- Record: found
- Abstract: found
- Article: not found
Single-copy insertion of transgenes in Caenorhabditis elegans.
- Record: found
- Abstract: found
- Article: not found
A genome-scale resource for in vivo tag-based protein function exploration in C. elegans.
Mihail Sarov, John I. Murray, Kristin Schanze … (2012)
- Record: found
- Abstract: found
- Article: not found
Distinct requirements for somatic and germline expression of a generally expressed Caernorhabditis elegans gene.
A Fire, Heather Montgomery, Conor W. Kelly … (1997)