- Record: found
- Abstract: found
- Article: found
A Reporter Screen in a Human Haploid Cell Line Identifies CYLD as a Constitutive Inhibitor of NF-κB
Read this article at
Abstract
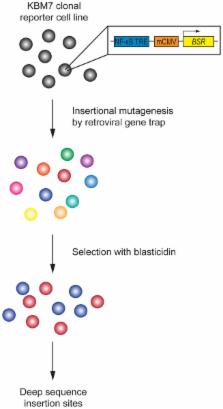
The development of forward genetic screens in human haploid cells has the potential to transform our understanding of the genetic basis of cellular processes unique to man. So far, this approach has been limited mostly to the identification of genes that mediate cell death in response to a lethal agent, likely due to the ease with which this phenotype can be observed. Here, we perform the first reporter screen in the near-haploid KBM7 cell line to identify constitutive inhibitors of NF-κB. CYLD was the only currently known negative regulator of NF-κB to be identified, thus uniquely distinguishing this gene. Also identified were three genes with no previous known connection to NF-κB. Our results demonstrate that reporter screens in haploid human cells can be applied to investigate the many complex signaling pathways that converge upon transcription factors.
Related collections
Most cited references13
- Record: found
- Abstract: found
- Article: not found
Haploid genetic screens in human cells identify host factors used by pathogens.
- Record: found
- Abstract: found
- Article: not found
The nuclear factor kappa B signaling pathway: integrating metabolism with inflammation.
- Record: found
- Abstract: found
- Article: not found