- Record: found
- Abstract: found
- Article: found
Sensing the environment by bacterial plant pathogens: What do their numerous chemoreceptors recognize?

Read this article at
Abstract
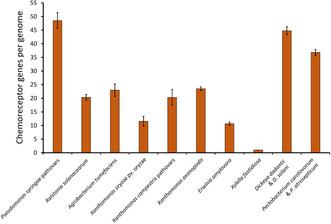
Bacteria have evolved multiple sensing strategies to efficiently adapt to their natural hosts and environments. In the context of plant pathology, chemotaxis allows phytopathogenic bacteria to direct their movement towards hosts through the detection of a landscape of plant‐derived molecules, facilitating the initiation of the infective process. The importance of chemotaxis for the lifestyle of phytopathogens is also reflected in the fact that they have, on average, twice as many chemoreceptors as bacteria that do not interact with plants. Paradoxically, the knowledge about the function of plant pathogen chemoreceptors is scarce. Notably, many of these receptors seem to be specific to plant‐interacting bacteria, suggesting that they may recognize plant‐specific compounds. Here, we highlight the need to advance our knowledge of phytopathogen chemoreceptor function, which may serve as a base for the development of anti‐infective therapies for the control of phytopathogens.
Abstract
Related collections
Most cited references62
- Record: found
- Abstract: found
- Article: not found
Top 10 plant pathogenic bacteria in molecular plant pathology.
- Record: found
- Abstract: not found
- Article: not found
Root exudates: from plant to rhizosphere and beyond
- Record: found
- Abstract: found
- Article: not found