- Record: found
- Abstract: found
- Article: found
A Genome-Wide mQTL Analysis in Human Adipose Tissue Identifies Genetic Variants Associated with DNA Methylation, Gene Expression and Metabolic Traits
Read this article at
Abstract
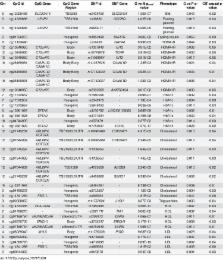
Little is known about the extent to which interactions between genetics and epigenetics may affect the risk of complex metabolic diseases and/or their intermediary phenotypes. We performed a genome-wide DNA methylation quantitative trait locus (mQTL) analysis in human adipose tissue of 119 men, where 592,794 single nucleotide polymorphisms (SNPs) were related to DNA methylation of 477,891 CpG sites, covering 99% of RefSeq genes. SNPs in significant mQTLs were further related to gene expression in adipose tissue and obesity related traits. We found 101,911 SNP-CpG pairs (mQTLs) in cis and 5,342 SNP-CpG pairs in trans showing significant associations between genotype and DNA methylation in adipose tissue after correction for multiple testing, where cis is defined as distance less than 500 kb between a SNP and CpG site. These mQTLs include reported obesity, lipid and type 2 diabetes loci, e.g. ADCY3/POMC, APOA5, CETP, FADS2, GCKR, SORT1 and LEPR. Significant mQTLs were overrepresented in intergenic regions meanwhile underrepresented in promoter regions and CpG islands. We further identified 635 SNPs in significant cis-mQTLs associated with expression of 86 genes in adipose tissue including CHRNA5, G6PC2, GPX7, RPL27A, THNSL2 and ZFP57. SNPs in significant mQTLs were also associated with body mass index (BMI), lipid traits and glucose and insulin levels in our study cohort and public available consortia data. Importantly, the Causal Inference Test (CIT) demonstrates how genetic variants mediate their effects on metabolic traits (e.g. BMI, cholesterol, high-density lipoprotein (HDL), hemoglobin A1c (HbA1c) and homeostatic model assessment of insulin resistance (HOMA-IR)) via altered DNA methylation in human adipose tissue. This study identifies genome-wide interactions between genetic and epigenetic variation in both cis and trans positions influencing gene expression in adipose tissue and in vivo (dys)metabolic traits associated with the development of obesity and diabetes.
Related collections
Most cited references33
- Record: found
- Abstract: found
- Article: not found
High density DNA methylation array with single CpG site resolution.
- Record: found
- Abstract: found
- Article: not found
Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis.
- Record: found
- Abstract: found
- Article: not found