- Record: found
- Abstract: found
- Article: found
A metabolomic approach to identifying platinum resistance in ovarian cancer
Read this article at
Abstract
Background
Acquisition of metabolic alterations has been shown to be essential for the unremitting growth of cancer, yet the relation of such alterations to chemosensitivity has not been investigated. In the present study our aim was to identify the metabolic alterations that are specifically associated with platinum resistance in ovarian cancer. A global metabolic analysis of the A2780 platinum-sensitive and its platinum-resistant derivative C200 ovarian cancer cell line was performed utilizing ultra-high performance liquid chromatography/mass spectroscopy and gas chromatography/mass spectroscopy. Per-metabolite comparisons were made between cell lines and an interpretive analysis was carried out using the Kyoto Encyclopedia of Genes and Genomes (KEGG) metabolic library and the Ingenuity exogenous molecule library.
Results
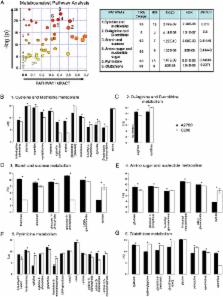
We observed 288 identified metabolites, of which 179 were found to be significantly different ( t-test p < 0.05) between A2780 and C200 cells. Of these, 70 had increased and 109 had decreased levels in platinum resistant C200 cells. The top altered KEGG pathways based on number or impact of alterations involved the cysteine and methionine metabolism. An Ingenuity Pathway Analysis also revealed that the methionine degradation super-pathway and cysteine biosynthesis are the top two canonical pathways affected. The highest scoring network of altered metabolites was related to carbohydrate metabolism, energy production, and small molecule biochemistry. Compilation of KEGG analysis and the common network molecules revealed methionine and associated pathways of glutathione synthesis and polyamine biosynthesis to be most significantly altered.
Conclusion
Our findings disclose that the chemoresistant C200 ovarian cancer cells have distinct metabolic alterations that may contribute to its platinum resistance. This distinct metabolic profile of platinum resistance is a first step towards biomarker development for the detection of chemoresistant disease and metabolism-based drug targets specific for chemoresistant tumors.
Related collections
Most cited references42

- Record: found
- Abstract: found
- Article: found
MetaboAnalyst: a web server for metabolomic data analysis and interpretation
- Record: found
- Abstract: found
- Article: not found
Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression.

- Record: found
- Abstract: found
- Article: found