- Record: found
- Abstract: found
- Article: found
Influence of N6-Methyladenosine Modification Gene HNRNPC on Cell Phenotype in Parkinson's Disease
Read this article at
Abstract
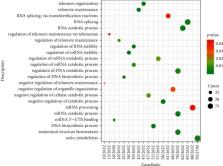
This study aimed to explore the N6-methyladenosine (m6A) modification genes involved in the pathogenesis of Parkinson's disease (PD) through data analysis of the two data sets GSE120306 and GSE22491 in the GEO database and further explore its influence on cell phenotype in PD. We analyzed the differentially expressed genes and function enrichment analysis of the two sets of data and found that the expression of the m6A-modification gene HNRNPC was significantly downregulated in the PD group, and it played an important role in DNA metabolism, RNA metabolism, and RNA processing and may be involved in PD. Then, we constructed the HNRNPC differential expression cell line to study the role of this gene in the pathogenesis of PD. The results showed that overexpression of HNRNPC can promote the proliferation of PC12 cells, inhibit their apoptosis, and inhibit the expression of inflammatory factors IFN- β, IL-6, and TNF- α, suggesting that HNRNPC may cause PD by inhibiting the proliferation of dopaminergic nerve cells, promoting their apoptosis, and causing immune inflammation. Our study also has certain limitations. For example, the data of the experimental group and the validation group come from different cell types, and the data of the experimental group involve individuals with G2019S LRRK2 mutations. In addition, due to the low expression of HNRNPC in PC12 cells, we used the method of overexpressing this gene to study its function. All these factors may cause our conclusions to be biased. Therefore, more research is still needed to corroborate it in the future.
Related collections
Most cited references53
- Record: found
- Abstract: found
- Article: not found
clusterProfiler: an R package for comparing biological themes among gene clusters.
- Record: found
- Abstract: found
- Article: not found
m6A-dependent regulation of messenger RNA stability
- Record: found
- Abstract: found
- Article: not found