- Record: found
- Abstract: found
- Article: found
Probing key elements of teixobactin–lipid II interactions in membranes†

Read this article at
Abstract
Abstract
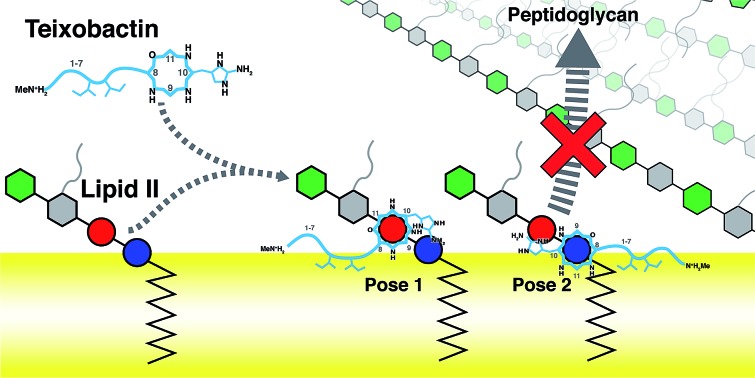
Teixobactin (Txb) is a recently discovered antibiotic against Gram-positive bacteria that induces no detectable resistance. The bactericidal mechanism is believed to be the inhibition of cell wall biosynthesis by Txb binding to lipid II and lipid III. Txb binding specificity likely arises from targeting of the shared lipid component, the pyrophosphate moiety. Despite synthesis and functional assessment of numerous chemical analogs of Txb, and consequent identification of the Txb pharmacophore, the detailed structural information of Txb–substrate binding is still lacking. Here, we use molecular modeling and microsecond-scale molecular dynamics simulations to capture the formation of Txb–lipid II complexes at a membrane surface. Two dominant binding conformations were observed, both showing characteristic lipid II phosphate binding by the Txb backbone amides near the C-terminal cyclodepsipeptide ( d-Thr8–Ile11) ring. Additionally, binding by Txb also involved the side chain hydroxyl group of Ser7, as well as a secondary phosphate binding provided by the side chain of l- allo-enduracididine. Interestingly, those conformations differ by swapping two groups of hydrogen bond donors that coordinate the two phosphate moieties of lipid II, resulting in opposite orientations of lipid II binding. In addition, residues d- allo-Ile5 and Ile6 serve as the membrane anchors in both Txb conformations, regardless of the detailed phosphate binding interactions near the cyclodepsipeptide ring. The role of hydrophobic residues in Txb activity is primarily for its membrane insertion, and subsidiarily to provide non-polar interactions with the lipid II tail. Based on the Txb–lipid II interactions captured in their complexes, as well as their partitioning depths into the membrane, we propose that the bactericidal mechanism of Txb is to arrest cell wall synthesis by selectively inhibiting the transglycosylation of peptidoglycan, while possibly leaving the transpeptidation step unaffected. The observed “pyrophosphate caging” mechanism of lipid II inhibition appears to be similar to some lantibiotics, but different from that of vancomycin or bacitracin.
Related collections
Most cited references3
- Record: found
- Abstract: found
- Article: not found
CHARMM additive all-atom force field for carbohydrate derivatives and its utility in polysaccharide and carbohydrate-protein modeling.
- Record: found
- Abstract: not found
- Article: not found
Hydrogen bonding in globular proteins
- Record: found
- Abstract: found
- Article: not found
The nisin-lipid II complex reveals a pyrophosphate cage that provides a blueprint for novel antibiotics.
Author and article information
Notes
†Electronic supplementary information (ESI) available: Computational methods and supplementary figures. See DOI: 10.1039/c8sc02616e