- Record: found
- Abstract: found
- Article: found
Text-based phenotypic profiles incorporating biochemical phenotypes of inborn errors of metabolism improve phenomics-based diagnosis

Read this article at
Abstract
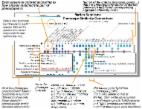
Phenomics is the comprehensive study of phenotypes at every level of biology: from metabolites to organisms. With high throughput technologies increasing the scope of biological discoveries, the field of phenomics has been developing rapid and precise methods to collect, catalog, and analyze phenotypes. Such methods have allowed phenotypic data to be widely used in medical applications, from assisting clinical diagnoses to prioritizing genomic diagnoses. To channel the benefits of phenomics into the field of inborn errors of metabolism (IEM), we have recently launched IEMbase, an expert-curated knowledgebase of IEM and their disease-characterizing phenotypes. While our efforts with IEMbase have realized benefits, taking full advantage of phenomics requires a comprehensive curation of IEM phenotypes in core phenomics projects, which is dependent upon contributions from the IEM clinical and research community. Here, we assess the inclusion of IEM biochemical phenotypes in a core phenomics project, the Human Phenotype Ontology. We then demonstrate the utility of biochemical phenotypes using a text-based phenomics method to predict gene-disease relationships, showing that the prediction of IEM genes is significantly better using biochemical rather than clinical profiles. The findings herein provide a motivating goal for the IEM community to expand the computationally accessible descriptions of biochemical phenotypes associated with IEM in phenomics resources.
Related collections
Most cited references8
- Record: found
- Abstract: found
- Article: found
The Monarch Initiative: an integrative data and analytic platform connecting phenotypes to genotypes across species
- Record: found
- Abstract: found
- Article: not found
Next-generation sequencing demands next-generation phenotyping.
- Record: found
- Abstract: not found
- Article: not found