- Record: found
- Abstract: found
- Article: found
Characterization and applications of Nanobodies against human procalcitonin selected from a novel naïve Nanobody phage display library
Read this article at
Abstract
Background
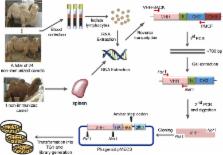
Nanobodies (Nbs) are single-domain antigen-binding fragments derived from the camelids heavy-chain only antibodies (HCAbs). Their unique advantageous properties make Nbs highly attractive in various applications. The general approach to obtain Nbs is to isolate them from immune libraries by phage display technology. However, it is unfeasible when the antigens are toxic, lethal, transmissible or of low immunogenicity. Naïve libraries could be an alternative way to solve the above problems.
Results
We constructed a large camel naïve phage display Nanobody (Nb) library with great diversity. The generated library contains to 6.86 × 10 11 clones and to our best of knowledge, this is the biggest naïve phage display Nb library. Then Nbs against human procalcitonin (PCT) were isolated from this library. These Nbs showed comparable affinity and antigen-binding thermostability at 37°C and 60°C compared to the PCT Nbs from an immune phage-displayed library. Furthermore, two PCT Nbs that recognize unique epitopes on PCT have been successfully applied to develop a sandwich enzyme-linked immunosorbent assay (ELISA) to detect PCT, which showed a linear working range from 10-1000 ng/mL of PCT.
Conclusion
We have constructed a large and diverse naïve phage display Nb library, which potentially functioning as a good resource for selecting antigen-binders with high quality. Moreover, functional Nbs against PCT were successfully characterized and applied, providing great values on medical application.
Related collections
Most cited references23
- Record: found
- Abstract: found
- Article: not found
A new antigen receptor gene family that undergoes rearrangement and extensive somatic diversification in sharks.
- Record: found
- Abstract: found
- Article: not found
Isolation of high affinity human antibodies directly from large synthetic repertoires.
- Record: found
- Abstract: found
- Article: not found