- Record: found
- Abstract: found
- Article: found
Deep connections: Divergence histories with gene flow in mesophotic Agaricia corals
Read this article at
Abstract
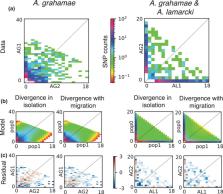
Largely understudied, mesophotic coral ecosystems lie below shallow reefs (at >30 m depth) and comprise ecologically distinct communities. Brooding reproductive modes appear to predominate among mesophotic‐specialist corals and may limit genetic connectivity among populations. Using reduced representation genomic sequencing, we assessed spatial population genetic structure at 50 m depth in an ecologically important mesophotic‐specialist species Agaricia grahamae, among locations in the Southern Caribbean. We also tested for hybridisation with the closely related (but depth‐generalist) species Agaricia lamarcki, within their sympatric depth zone (50 m). In contrast to our expectations, no spatial genetic structure was detected between the reefs of Curaçao and Bonaire (~40 km apart) within A. grahamae. However, cryptic taxa were discovered within both taxonomic species, with those in A. lamarcki (incompletely) partitioned by depth and those in A. grahamae occurring sympatrically (at the same depth). Hybrid analyses and demographic modelling identified contemporary and historical gene flow among cryptic taxa, both within and between A. grahamae and A. lamarcki. These results (1) indicate that spatial connectivity and subsequent replenishment may be possible between islands of moderate geographic distances for A. grahamae, an ecologically important mesophotic species, (2) that cryptic taxa occur in the mesophotic zone and environmental selection along shallow to mesophotic depth gradients may drive divergence in depth‐generalists such as A. lamarcki, and (3) highlight that gene flow links taxa within this relativity diverse Caribbean genus.
Related collections
Most cited references114

- Record: found
- Abstract: found
- Article: found
The variant call format and VCFtools
- Record: found
- Abstract: found
- Article: not found
Detecting the number of clusters of individuals using the software structure: a simulation study
- Record: found
- Abstract: found
- Article: not found