- Record: found
- Abstract: found
- Article: not found
The Genetic Program of Pancreatic β-Cell Replication In Vivo
Read this article at
Abstract
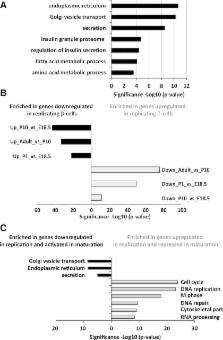
The molecular program underlying infrequent replication of pancreatic β-cells remains largely inaccessible. Using transgenic mice expressing green fluorescent protein in cycling cells, we sorted live, replicating β-cells and determined their transcriptome. Replicating β-cells upregulate hundreds of proliferation-related genes, along with many novel putative cell cycle components. Strikingly, genes involved in β-cell functions, namely, glucose sensing and insulin secretion, were repressed. Further studies using single-molecule RNA in situ hybridization revealed that in fact, replicating β-cells double the amount of RNA for most genes, but this upregulation excludes genes involved in β-cell function. These data suggest that the quiescence-proliferation transition involves global amplification of gene expression, except for a subset of tissue-specific genes, which are “left behind” and whose relative mRNA amount decreases. Our work provides a unique resource for the study of replicating β-cells in vivo.
Related collections
Most cited references31
- Record: found
- Abstract: found
- Article: not found
Revisiting global gene expression analysis.
- Record: found
- Abstract: found
- Article: not found
Selective transcriptional regulation by Myc in cellular growth control and lymphomagenesis.
- Record: found
- Abstract: found
- Article: not found
