INTRODUCTION
It is well known that RNA secondary structures are vitally important for the life cycle of many DNA and RNA containing viruses. RNA hairpins are required for viral replication, mRNA editing, microRNA biogenesis, splicing, genome packaging, etc. [1, 2]. Internal ribosome entry sites (IRES), consisting of extensive regions of structured RNA, are key elements for the initiation of cap-independent translation and have been found in many pathogenic viruses [3–6]. RNA secondary structures of Rous sarcoma virus, retroviruses, coronaviruses, and many other viruses are responsible for switching between the expression of overlapping reading frames, which expand the information capacity of the viral genome [7, 8]. Hairpin-type secondary structures of flavivirus genomes provide the translation of full-length capsid protein in the presence of multiple start codons and are required for viral replication [9]. Conserved corkscrew structures in terminal regions have been found in various groups of negative-sense RNA viruses, including influenza, and are involved in various steps of viral replication [10].
The influenza A virus (IAV) genome consists of eight segments of negative-sense RNA that encode up to 18 proteins [11]. During the process of viral replication, positive-sense RNA is also synthesized, which is classified into a genomic complementary (+)RNA (cRNA) or messenger RNA (mRNA). Currently, the function of several secondary structures within promoter regions of vRNA and cRNA, and splice sites in the M and NS genes [12] are only partially understood.
The most precise secondary structure of (+)RNA has been determined for the NS segment of influenza A virus [13–17]. This genomic segment encodes at least two important non-structural proteins (NS and NEP), and contains a splicing regulated switch between the corresponding open reading frames (ORFs). The multifunctional NS1 protein is involved in the inhibition of the cellular immune response during viral infection [18]. NS1 expression varies among IAV strains. For example, A/Brevig Mission/1/1918 (H1N1), responsible for the largest pandemic in human history, and considered the ancestor of all modern human IAV strains, has a high level of NS1 expression compared to less pathogenic strains [19]. The main function of the NEP protein is related to the nuclear export of viral ribonucleoproteins [20]. The optimal expression of NEP protein contributes to the maintenance of effective viral replication [21–23].
Regions with a conserved secondary structure, corresponding to NS (+)RNA position 82-148 and position 497-564, are localized close to the 5’ and 3’ splice sites. The first region is located within the NS1 ORF; the second region is located within NS1 and NEP ORFs [24]. According to previously published data, both NS (+)RNA regions fold into several alternative secondary structures: region 82-148 forms hairpin or multi-branch structures [15–17]; region 497-564 is characterized by the formation of two structures with similar energies (pseudoknot or hairpin) [14].
It is noteworthy that NS (+)RNA secondary structures differ among IAV strains [14, 24, 25]. The highly virulent A/Brevig Mission/1/1918 (H1N1) strain has stable RNA hairpin structures in both of the aforementioned (NS RNA) regions. Highly virulent avian influenza A (H5N1) strains that appeared after 2001 carry more stable RNA hairpins in the second region compared to previous strains [14]. Since the NS gene is closely associated with host adaptation, structural motifs may affect host specificity or viral pathogenicity through the regulation of NS1 expression [15, 26]. However, the role of the observed hairpins is yet to be determined.
Here, we present the study results of 4 previously constructed viruses based on the A/Puerto Rico/8/1934 (H1N1) strain, featuring different combinations of secondary structures in the NS mRNA region 82-148 and region 497-564 [26]. The NS gene sequences, providing stable RNA hairpins in the corresponding regions, were adopted from the aforementioned influenza strains, while the sequences with no stable secondary structures were taken from their closest “counterparts” with a minimum number of mismatches (Fig. 1). RNA secondary structure analysis was performed using the RNAfold online tool and did not include a pseudoknot structure prediction [27]. The difference in the NS region 82-148 mRNA structure was concurrent with 2 synonymous mutations (G123A, A132G). The difference in the NS region 497-564 mRNA structure was concurrent with one non-synonymous substitution in the NS1 open reading frame (G166N) and two non-synonymous substitutions in the NEP ORF (M14V, G22A). Virus variants bearing different secondary structure combinations were named according to the presence (1) or absence (0) of an RNA hairpin in the NS region 82-148 and/or region 497-564. The variants were named as: V1-1, V1-0, V0-1, or V0-0.

MATERIALS AND METHODS
Viruses
Four virus variants with different combinations of secondary structures in the two NS RNA regions (82-148 and 497-564) were previously obtained by reverse genetics, followed by the virus production in Vero cells [26]. All four variants are based on the laboratory strain A/Puerto Rico/8/1934 (H1N1).
Cell culture
Madin-Darby Canine Kidney (MDCK) London line cells (International Reagent Resource, #FR-58) were grown in AlphaMEM medium (BioloT, Russia) supplemented with 2 mM L-glutamine and 10% SC-biol FBS (BioloT, Russia). A549 cells (ATCC, #CCL-185) were cultivated in DMEM/ F12 media (Gibco) supplemented with 2% GlutaMAX (Gibco) and 10% FBS (Gibco). The Vero cell line (ATCC, #CCL-81) was previously adapted to an OptiPro serum-free medium (Gibco) supplemented with 2% GlutaMAX (Gibco). Primary chicken kidney cells culture (CK, kindly provided by K. A. Vasiliev, Laboratory of Vector Vaccines, Smorodintsev Research Institute of Influenza) were grown in a DMEM/F12 media (Gibco) supplemented with 1% GlutaMAX (Gibco), 1% sodium pyruvate (Gibco), and 10% SC-biol FBS (BioloT, Russia).
Viral infectious activity
Infectious activity of virus samples was measured by titration in cells (overnight confluent, 96-well plate format) using serum-free culture media (described above) supplemented with 1% antibiotic-antimycotic (Gibco) and TPCK-trypsin (Sigma). Trypsin concentrations were: 2.5 μg/ml for MDCK; 0.5 μg/ml for A549; and 0.5 μg/ml for Vero cells. Titration in primary CK cells was performed without trypsin. Titration in 10-day old embryonated chicken eggs was conducted by the inoculation of the allantoic cavity with 200 μl of 10-fold diluted virus sample in PBS supplemented with 1% antibiotic-antimycotic (Gibco). The 50% Tissue Culture Infectious Dose (TCID50) and the 50% Embryonic Infectious Dose (EID50) were calculated according to the Reed and Muench method [28] following the hemagglutination assay (HA) performed with 0.75% suspension of chicken red blood cells. Viral infectious activity was expressed as log10TCID50/ml or log10EID50/ml.
Western blotting
A549 cells (overnight confluent monolayer, 12-well plate format, in triplicate) were infected with each virus at a multiplicity of infection (moi) of 1 TCID50/cell. After 1 h of virus adsorption at 37°C, inoculum was replaced with serum-free medium. After 9-12 h of incubation, the medium was removed. Cells were then scraped, washed once, and resuspended in 100 μl PBS. Sample protein concentrations were measured using the DC Protein Assay Kit I (Bio-Rad, #5000111) in triplicates. Then, samples (0.5 μg or 15 μg of total protein for NS1 or NEP detection, respectively) were mixed with 1X Laemmli Sample Buffer (Bio-Rad) containing β-mercaptoethanol and incubated for 5 min. at 100°C. The samples were separated by SDS PAGE using Mini-PROTEAN TGX Stain-Free Precast Protein Gels (Bio-Rad, #4568121) following the manufacturer’s instructions.
After separation, proteins were transferred onto nitrocellulose membranes using the Trans-Blot Turbo Mini (Bio-Rad, #1704156). Membranes were blocked overnight at 4°C in 5% low fat milk (Bio-Rad) in PBST (PBS with 0.1% tween-20), followed by 1 h of incubation with primary antibody at room temperature. We used the following antibodies (1 μg/ml in blocking buffer): mouse monoclonal antibody 1H7 against NS1 [29]; rabbit monoclonal antibody against NEP (Novus Biologicals #NBP2-42872); and mouse monoclonal antibody against alphatubulin (Sigma-Aldrich #T6074). The membranes were washed twice in PBST and incubated with GAM-HRP (Bio-Rad, #1706515) or GAR-HRP (Bio-Rad #1721011) secondary antibodies (1 μg/ml in blocking buffer) for 1 h at room temperature. After two washes with PBST, the membranes were incubated with the Clarity Western ECL Substrate kit (Bio-Rad) and analyzed with the ChemiDoc System (Bio-Rad). The ImageLab 4.1 (Bio-Rad) and ImageJ 1.38 [30] programs were used for the quantitative analysis.
Laboratory animal infection
Female BALB/c mice (18–20 g, aged 6-8 weeks) were purchased from the Stolbovaya Biomedical Scientific Center FMBA (Russia). Animal care and all of the experiments were performed according to international guidelines [31]. Animals were randomly divided into four groups and intranasally infected with viruses diluted in DPBS (Biolot, Russia) (30 μl/mouse) under mild ether anesthesia. After infection, animals were monitored daily, including the weighing and assessment of mortality. The testing for the presence of virus-specific antibodies in the blood sera of surviving mice was performed on day 21 after infection using the standard hemagglutination inhibition (HI) assay. The calculation of the 50% mouse lethal dose (MLD50) and 50% mouse infectious dose (MID50) were performed according to the Reed and Muench method [28].
In order to estimate the viral replication in animal lungs, the mice were infected with 10 MID50 of each virus. Four mice from each group were euthanized at specific time points (2, 4, and 6 days post infection (pi)). The lungs were isolated, and 10% tissue suspensions were prepared in DPBS (Biolot, Russia) with 1% antibiotic-antimycotic (Gibco). After centrifugation (3000 rpm, 10 min), suspensions were used to determine the presence of infectious viral particles by titration in MDCK cell culture.
Statistics
Statistical data analysis was performed using Microsoft Excel 2007 and GraphPad Prism 6.01. Two-way ANOVA with Tukey’s or Sidak’s post-test was used to assess the significance of any differences between viruses at specific time points. The significance threshold was set at p<0.05. The data are presented as mean ± standard deviation (M±SD).
RESULTS
Infectious activity of viruses in different living systems
Four influenza viruses (V1-1, V1-0, V0-1, V0-0), based on the A/Puerto Rico/8/34 strain and with different combinations of NS (+)RNA secondary structure in the region 82-148 and region 497-564, were previously constructed [26]. We first compared the infectious activity of the viruses in Vero, MDCK, A549, and CK cells and in 10-day old embryonated chicken eggs. The results shown in Table 1 indicate that there were no statistically significant differences between the viruses, indicating equal amounts of infectious viral particles in each sample.
| Cells | Infectious activity (log10TCID50/ml or log10EID50/ml) | |||
|---|---|---|---|---|
| V1-1 | V1-0 | V0-1 | V0-0 | |
| A549 | 7.4±0.55 | 7.78±0.69 | 7.94±0.59 | 7.91±0.64 |
| MDCK | 9.44±0.10 | 9.89±0.18 | 9.17±0.29 | 9.19±0.50 |
| Vero | 8.78±0.19 | 8.56±0.10 | 8.72±0.25 | 8.81±0.34 |
| CK | 7.55±0.11 | 7.95±0.48 | 7.56±0.10 | 7.56±0.10 |
| CE | 9.89±0.97 | 9.47±0.42 | 9.22±0.29 | 9.53±0.29 |
Data are presented as M±SD (n=3)
First cycle viral growth kinetics in vitro
In previous research, we evaluated the replication ability of viruses by multi-cycle growth curves in Vero, MDCK, or A549 cells [26]. We did not observe any significant differences in multi-cycle cell infection. However, the viruses differed from each other in infectious activity 48h post transfection. This suggests that the aforementioned RNA structures may be important in the early stages of infection [26]. Over the course of this project, we studied the replication properties of the viruses with different NS mRNA secondary structures during the first viral cycle (Fig. 2).
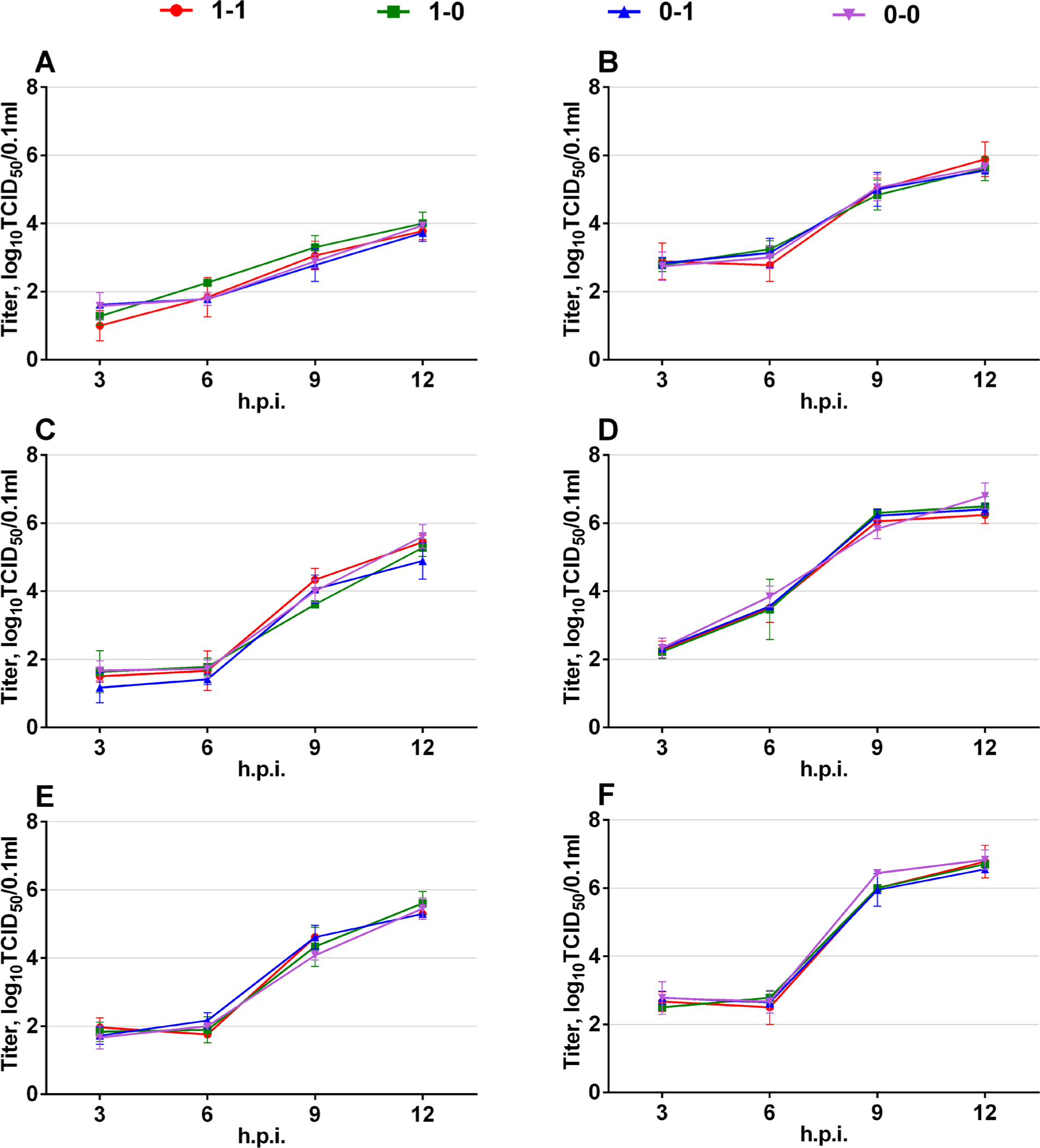
A549 (A, B), MDCK (C, D), and Vero (E, F) cells were infected at a moi of 0.1 or 1 TCID50/cell. Infectious activity of viral progeny was measured at the indicated time points by TCID50 assay. Data are presented as M ± SD (n=3).
A549, MDCK, and Vero cells were infected with each virus at a MOI of 0.1 or 1.0 (TCID50/cell) in triplicates (overnight confluent, 6-well plate format). At 3, 6, 9, and 12 hours pi (hpi), viral progeny was collected from the supernatants, and infectious titers were measured in MDCK cells. According to the obtained results, no differences were found between the strains. Therefore, there was no correlation between viral replication in cell culture and the types of NS (+)RNA secondary structure present.
Viral protein production levels in infected cells
In order to estimate how the RNA structure affects viral protein expression, the content of NEP and NS1 proteins in infected A549 whole cell lysate was measured by Western blot at 9 and 12 hpi, respectively (Fig. 3). The relative expression levels were determined by measuring the intensities of the obtained bands with subsequent normalization to that of α-tubulin. It was found that NEP levels in the cells infected with V0-0 were higher than those of other samples. The lowest NEP production corresponded to V0-1. Viruses V1-1 and V1-0 had comparable levels of NEP (Fig. 3A).
Western blot also confirmed that the hairpin in the NS (+)RNA region 82-148 enhanced NS1 expression during early infection in A549 cells (Fig. 3B). According to measurements, the production level of NS1 in cells infected with V1-1 or V1-0 was approximately twice higher than that seen in V0-1 or V0-0 infected cells which is entirely consistent with previously published results obtained by ELISA [26].
Therefore, NS (+)RNA secondary structures were found to affect the production of viral proteins in infected cells. Despite the fact that NS (+)RNA secondary structures did not affect viral replication in vitro, the study was continued using an in vivo model. This approach was followed because active NS1 protein synthesis in the first hours of infection may inhibit the interferon (IFN) dependent immune response and thereby affect disease severity.
Viral reproduction in infected mice
There were no statistically significant differences between the four viral variants in terms of MID50 and MLD50. The standard deviations of MLD50 and MID50 were within +/-0.5 log10TCID50, which is within the error of the titration method (Table 2). To study the dynamics of the (viral) infectious process, animals were infected with each virus at a dose of 10 MID50 per mouse. They were monitored daily, and the presence of virus in the lungs was determined at 2, 4, and 6 days pi. According to the data, V0-1 was less pathogenic for mice. Mice infected with V0-1 had a tendency to lose weight more slowly compared to other animals (Fig. 4A). On the 2nd day, a difference between V1-1 and V0-1 viruses was observed. On the 6th day after infection, a decrease in lung infectious titer of V0-1 was seen (Fig. 4B). This proves that a specific combination of NS (+)RNA secondary structures affects viral pathogenicity in mice.
| Virus | 1 MLD50 (log10TCID50/ml) | 1 MID50 (log10TCID50/ml) |
|---|---|---|
| V1-1 | 3.43±0.18 | 1.95±0.50 |
| V1-0 | 3.09±0.65 | 1.65±1.01 |
| V0-1 | 3.59±1.36 | 1.12±0.17 |
| V0-0 | 2.68±0.53 | 1.05±0.21 |
Data are presented as (M ± SD) for two independent experiments.

Mice were infected with 10 MID50 of each virus. (A) The dynamics of the weight of infected animals measured individually and presented as mean % of initial weight of the group; (B) Viral replication in lungs of infected mice. The viral infectious activity was measured by TCID50 assay of 10% lung suspensions. Individual values are presented. Sign (*) indicates the significant differences between V1-1 and V0-1 on day 2, p<0.05; significant differences between V0-1 and other viruses on day 6, p<0.05 (two-way ANOVA with Sidak’s multiple comparisons test).
DISCUSSION
Previously published experiments devoted to NS gene secondary structures were performed only in cell culture [32] or in vitro [15] without taking into account the possible contributions of the structural combinations. In this study, we have shown that certain NS (+)RNA secondary structure combinations regulate NS1 and NEP production in vitro and can affect the level of viral pathogenicity in vivo.
Here we have demonstrated, for the first time, that a specific combination (presence of a hairpin at position 497-564 and absence of a hairpin structure at position 82-148) leads to decreased NEP expression in infected A549 cells (V0-1 compared to V0-0) (Fig. 3A).
If RNA hairpin was present in 82-148 region, there was no effect of RNA secondary structure in the second region (497-564) on the NEP expression. The V1-1 and V1-0 variants displayed similar NEP expression levels (Fig.3).
Despite the fact that the region 497-564 falls in the NS1 ORF, the hairpin status there did not affect NS1 protein expression, regardless of the region 82-148 structural status. This is best seen in Fig.3B when comparing: V0-1 with V0-0; and V1-1 with V1-0.
In contrast, the region 82-148 RNA hairpin drastically increased NS1 expression, regardless of the region 497-564 structural status (comparison of V1-1 with V0-1; V1-0 with V0-0). Variations in the NS1 expression level were previously demonstrated by ELISA [26] and additionally confirmed in this study using Western blot. Viral construct V1-0 (bearing a single RNA hairpin structure, from the A/Brevig Mission/1/1918 (H1N1) strain, in the region 82-148 of NS (+)RNA) was characterized by higher NS1 expression and lower NEP expression in infected A549 cells relative to a V0-0, which bears no hairpin in either region (Fig. 3). Since region 82-148 is only included in the NS1 ORF, the negative effect on NEP production probably occurs indirectly.
NS1 mRNA and NEP mRNA share the first 56 nucleotides including the untranslated region (5′-UTR) and first 30 nucleotides of the coding sequence. However, the NS1 mRNA in V1- 0 bears hairpin 82-148, presumably providing a more efficient ribosome-NS1 mRNA interaction, leading to high NS1 expression. This may lead to a lack of ribosome availability for NEP mRNA, which is approximately tenfold less present in infected cells compared to the unspliced NS mRNA form [33,34]. Therefore, we observed a negative influence of this structure on NEP expression and the suppression of region 497-564 structural effects on NEP translation. However, the mechanisms by which these secondary structure combinations affect NEP expression require further clarification.
Earlier, the stabilization of the RNA structure in the region 497-564 was demonstrated to change NS mRNA splicing in cell culture [32]. However, we did not observe the aforementioned difference between strains in our previous study measuring NS1 and NEP mRNA levels [26]. Therefore, we assume that NS (+)RNA secondary structures have effects at the level of translation: the hairpin in the region 82-148 enhances NS1 production, while the hairpin in region 497-564 inhibits NEP expression.
Our current in vivo viral replication experiment showed differences between the strains, depending on NS (+)RNA secondary structure combinations. Animals infected with V0-1 (without a hairpin in region 82-148 and a stable hairpin in region 497-564) contained less viral load in the lungs compared to animals infected with the other virus variants. This suggests that V0-1 is less pathogenic than the other strains. We assume that the absence of an RNA hairpin in the region 82-148 causes decreased NS1 protein expression, leading to a reduced ability of V0-1 to resist the IFN dependent immune response compared to the V1-1. The presence of an RNA hairpin in the region 497-564 results in decreased NEP protein expression, which apparently can also cause the mild attenuation of V0-1 in laboratory animals. The NEP protein controls the optimal speed of the viral cycle and, therefore, its decreased expression can negatively affect the viral replication process [35].
It should be mentioned that the different RNA secondary structures (between the A/Puerto Rico/8/1934-derived virus variants used in this study) were obtained by introducing mutations into the aforementioned RNA regions. The mutations (G511A, G512A, C537G) responsible for the secondary structure changes in the RNA region 497-564 cause one amino acid substitution in NS1 (N166G) and two substitutions in NEP (V14M and A22G). Amino acid position 166 is located in the proline-rich 162-170 loop of NS1 and is not included in the known structural or functional domains of the NS1 protein [36]. Therefore, we think it is unlikely that this substitution significantly contributes to the observed differences between virus variants, although this cannot be ruled out.
The NEP amino acid substitutions are located in the mobile, unstructured region of the N-terminal domain, the flexibility of which may be important for interaction with cellular proteins during nuclear export of viral RNA [37]. The aforementioned amino acids (valine/methionine at position 14, glycine/alanine at position 22) do not differ by hydrophobicity or charge. NEP amino acid residue 14 belongs to the 12-21 nuclear export signal, while position 22 does not affect any functional sequences. Therefore, V14M amino acid substitution may influence the viral RNA export process, leading to viral protein expression differences between pairs of viruses (V1-1 and V1-0; V0- 1 and V0-0). Since we did not observe the effect of the indicated mutations on the NS1 or NP expression in cell culture (results not shown), we suggest that their impact on the viral RNA export process is not significant. Nevertheless, the contribution of non-synonymous mutations to the observed phenotypes cannot be completely excluded from consideration.
It is important to note that our data contradicts an earlier hypothesis asserting that a hairpin in the region 497-564, typically found in highly virulent H5N1 strains, may act as an additional pathogenicity factor [14]. The formation of a stable RNA hairpin in such viruses may be host-specific, associated with higher body temperatures in birds, or an additional secondary structure stabilization requirement (compared to human strains) [13, 38].
Overall, we have demonstrated that NS mRNA secondary structures can affect viral replication in vivo. Previously, it was pointed out that regions of viral RNA featuring conserved secondary structures may be used as potential targets in new generations of antiviral therapy [12, 39] or for the development of live vaccines [32]. However, considering the slight contribution of NS mRNA secondary structures to viral pathogenesis, these structures are unlikely to be significant pathogenicity factors, but are rather involved in the fine regulations of NS1 and NEP gene expression.
