INTRODUCTION
Influenza viruses are enveloped viruses with a negative-sense single-stranded segmented RNA genome that belong to the Orthomyxoviridae family [1]. This family includes four genera namely influenza A virus (IAV), influenza B virus (IBV), influenza C virus (ICV), and influenza D virus (IDV) [2]. IAV infects a wide range of hosts including humans and causes seasonal epidemics and occasional pandemics in the human population, which is of great concern with respect to human health and economic well-being [3]. The genome of IAV consists of eight genomic segments.
Nuclear export protein (NEP) (also known as non-structural protein 2, NS2) with a sequence length of 121 amino acids, is encoded by the eighth segment via alternative splicing. NEP exhibits a highly conserved amino acid sequence and plays a key role in the infectious life cycle of the influenza virus by participating in the export of viral ribonucleoprotein (vRNP) complexes from the nucleus to the cell membrane, where they are assembled into virion progeny (see Fig. 1) [4, 5]. NEP also facilitates viral budding via interaction with ATPase [6]. During the process of the transcription and translation of viral RNA, NEP regulates the accumulation of viral RNA [7].
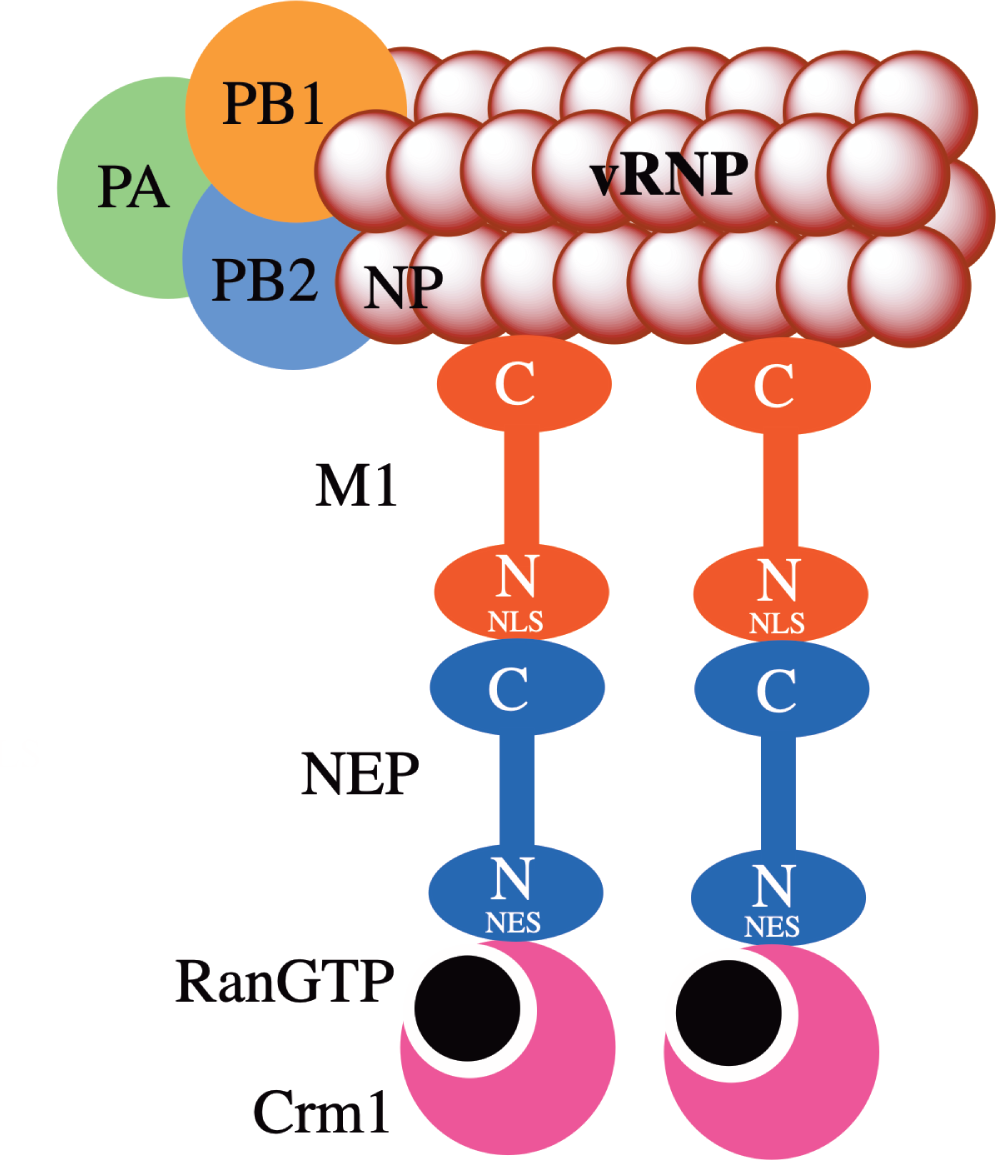
IAV NEP comprises two domains, namely N-terminal domain (residues 1-54), which is also known as the nuclear export signal (NES) domain, and C-terminal domain (residues 63-121), which is known as M1 binding domain [8]. The N-terminal domain consists of two NESs: NES1 (residues 12-21) and NES2 (residues 31-40) [9, 10]. The C-terminal domain interacts with the nuclear localization signal (NLS) of M1 protein via exposed Trp 78 residue surrounded by a cluster of negatively charged Glu residues [11]. This interaction is critical for the export of newly synthesized viral ribonucleoproteins from the host cell nucleus to the cytoplasm [11]. The complete X-ray structure of IAV NEP has eluded researchers to date. Only the structure of the C-terminal domain of NEP has been experimentally determined [11]. The N-terminal domain of NEP adopts highly flexible and mobile confirmation that contributes to the plasticity of the NEP [12]. The C-terminal domain consists of two alpha helices that form a dimer [11].
Ivermectin is a broad-spectrum antiparasitic drug that is widely used in veterinary medicine due to its safety and efficacy [13]. It has previously been shown that this drug has antiviral action against a broad variety of viruses. Ivermectin has been shown to act as an inhibitor of importin α/β-mediated nuclear import [14]. Ivermectin was also shown to inhibit human immunodeficiency virus (HIV-1) integrase nuclear import [14], import of the large tumor antigen of simian virus 40 [15], import of dengue virus non-structural protein 5 [14], and blocks the nuclear localization signal of parvovirus [16]. Ivermectin has been shown to impede infection caused by dengue virus 1-4 [17], West Nile Virus [18], Venezuelan equine encephalitis virus [19], influenza virus [20], pseudorabies virus [21], and severe acute respiratory syndrome coronavirus 2 [22].
Recently, four potential binding sites of ivermectin were found on the M1 binding domain (C-terminal domain) of IAV NEP via a search for structural fragments that are similar to the structure of ivermectin binding sites of different proteins using the information from the protein data bank (PDB) [23]. In the present study, we used in silico molecular docking to test the ability of ivermectin to bind to IAV NEP. We showed that ivermectin strongly binds to IAV NEP that should block the NEP-M1 protein interaction. It is anticipated that the blocking of this interaction can have a considerably deleterious effect on IAV assembly and propagation. Therefore, ivermectin could be used as a potential IAV NEP protein blocker in the treatment of influenza that would be another successful example of drug repurposing.
METHODS
IAV NEP and ivermectin structure retrieval
The protein structure of IAV NEP was retrieved from the protein data bank with the PDB identifier 1PD3 (https://www.rcsb.org/structure/1PD3) [11]. The obtained file (.pdb file) contained the structures of two chains – chain A and chain B. The chain B was removed, and the amino acid sequence 63-116 of IAV NEP that comprises chain A was analyzed. The ivermectin structure (.sdf file) was retrieved from PubChem (https://pubchem.ncbi.nlm.nih.gov/) with PubChem CID 6321424 [24].
Molecular docking experiment
The docking of the IAV NEP with ivermectin was carried out using the CB-Dock docking server (http://cao.labshare.cn/cb-dock/) [25]. CB-Dock is a protein-ligand docking server performing blind docking that involves four steps. In step 1, putative binding sites are detected on the protein structure. In step 2, several top binding sites are selected. The selection is based on the size of the binding sites. In step 3, the docking center is calculated and the docking box size is adjusted. In step 4, docking is carried out using AutoDock Vina and, after the completion of docking, binding sites are ranked according to the docking score [25].
RESULTS AND DISCUSSION
By providing an IAV NEP (.pdb file) and ivermectin structures (.sdf file) as an input to the CB-Dock server, a total of five binding sites for ivermectin were identified on NEP with varying Vina score. By default, the conformation with the highest score is treated as the best binding position and the corresponding site is considered as the optimal binding site for ivermectin [25]. Ivermectin was found to bind IAV NEP with a high AutoDock Vina score of -7.3 kcal/mol [cavity size: 185Å, center: 41*9*2 (x*y*z), and size: 29*29*29 (x*y*z)] (Fig. 2). Ivermectin interacts with the following amino acid residues at the binding site: Arg66, Glu67, Leu69, Gly70, Gln71, Glu74, Glu75, Arg77, Trp78, Leu107, Glu110, Gln111, Ile113, and Arg114. Of these, amino acids Arg66, Leu69, Glu74, Glu75, Trp78, Glu110, and Arg114 were found to be conserved [4]. Previously, four of ivermectin’s binding sites were identified on the C-terminal domain of NEP by means of the COFACTOR algorithm using the data from the protein data bank (PDB) [23]. Since these results are based on the structural similarity of ivermectin binding sites in different proteins with the structural fragments of the C-terminal domain of NEP and the authors of this study did not perform the in silico docking experiment to model the ivermectin binding to the predicted sites, these binding sites should be considered as hypothetical. According to the published data, Trp78 and the surrounding Glu residues as well as Glu81-Met100 in the M1 binding domain play a critical role in NEP-M1 protein interaction [11, 26]. The experiment described by Akarsu et al. [11] is based on the comparative study of NEP mutant proteins with one or several substitutions in amino acid sequence. Since Akarsu et al. proved that the folding of the mutant proteins corresponds to the folding of the wild type protein, their results are considered as the most reliable to date [6, 11]. The ivermectin binding site identified in this study comprises Trp78 residue as well as Glu67, Glu74, Glu75, and Glu110 residues. Since Trp78 along with the surrounding Glu residues was identified as the critical epitope for NEP binding to M1 protein [11], the NEP-M1 proteins association would be completely abolished by the interaction of IAV NEP with ivermectin because it binds to the same amino acid residues on the surface of NEP. That will prevent the export of newly synthesized viral ribonucleoproteins from the host nucleus to the cytoplasm which in turn will severely affect the life cycle of IAV. This makes ivermectin a promising anti-IAV drug molecule.


