- Record: found
- Abstract: found
- Article: found
Identification of a New Antimicrobial Resistance Gene Provides Fresh Insights Into Pleuromutilin Resistance in Brachyspira hyodysenteriae, Aetiological Agent of Swine Dysentery
Read this article at
Abstract
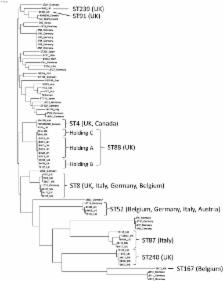
Brachyspira hyodysenteriae is the aetiological agent of swine dysentery, a globally distributed disease that causes profound economic loss, impedes the free trade and movement of animals, and has significant impact on pig health. Infection is generally treated with antibiotics of which pleuromutilins, such as tiamulin, are widely used for this purpose, but reports of resistance worldwide threaten continued effective control. In Brachyspira hyodysenteriae pleuromutilin resistance has been associated with mutations in chromosomal genes encoding ribosome-associated functions, however the dynamics of resistance acquisition are poorly understood, compromising stewardship efforts to preserve pleuromutilin effectiveness. In this study we undertook whole genome sequencing (WGS) and phenotypic susceptibility testing of 34 UK field isolates and 3 control strains to investigate pleuromutilin resistance in Brachyspira hyodysenteriae. Genome-wide association studies identified a new pleuromutilin resistance gene, tva(A) ( tiamulin valnemulin antibiotic resistance), encoding a predicted ABC-F transporter. In vitro culture of isolates in the presence of inhibitory or sub-inhibitory concentrations of tiamulin showed that tva(A) confers reduced pleuromutilin susceptibility that does not lead to clinical resistance but facilitates the development of higher-level resistance via mutations in genes encoding ribosome-associated functions. Genome sequencing of antibiotic-exposed isolates identified both new and previously described mutations in chromosomal genes associated with reduced pleuromutilin susceptibility, including the 23S rRNA gene and rplC, which encodes the L3 ribosomal protein. Interesting three antibiotic-exposed isolates harboured mutations in fusA, encoding Elongation Factor G, a gene not previously associated with pleuromutilin resistance. A longitudinal molecular epidemiological examination of two episodes of swine dysentery at the same farm indicated that tva(A) contributed to development of tiamulin resistance in vivo in a manner consistent with that seen experimentally in vitro. The in vitro studies further showed that tva(A) broadened the mutant selection window and raised the mutant prevention concentration above reported in vivo antibiotic concentrations obtained when administered at certain doses. We show how the identification and characterisation of tva(A), a new marker for pleuromutilin resistance, provides evidence to inform treatment regimes and reduce the development of resistance to this class of highly important antimicrobial agents.
Related collections
Most cited references44
- Record: found
- Abstract: found
- Article: not found
mcr-1 and mcr-2 variant genes identified in Moraxella species isolated from pigs in Great Britain from 2014 to 2015.
- Record: found
- Abstract: found
- Article: not found
A spreadsheet for the calculation of comprehensive statistics for the assessment of diagnostic tests and inter-rater agreement.
- Record: found
- Abstract: found
- Article: not found