- Record: found
- Abstract: found
- Article: found
Computational tools for the evaluation of laboratory-engineered biocatalysts
Read this article at
Abstract

Abstract
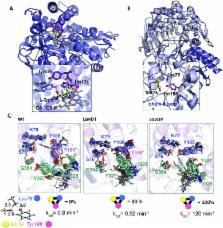
Biocatalysis is based on the application of natural catalysts for new purposes, for which enzymes were not designed. Although the first examples of biocatalysis were reported more than a century ago, biocatalysis was revolutionized after the discovery of an in vitro version of Darwinian evolution called Directed Evolution (DE). Despite the recent advances in the field, major challenges remain to be addressed. Currently, the best experimental approach consists of creating multiple mutations simultaneously while limiting the choices using statistical methods. Still, tens of thousands of variants need to be tested experimentally, and little information is available on how these mutations lead to enhanced enzyme proficiency. This review aims to provide a brief description of the available computational techniques to unveil the molecular basis of improved catalysis achieved by DE. An overview of the strengths and weaknesses of current computational strategies is explored with some recent representative examples. The understanding of how this powerful technique is able to obtain highly active variants is important for the future development of more robust computational methods to predict amino-acid changes needed for activity.
Related collections
Most cited references125
- Record: found
- Abstract: found
- Article: not found
Biomolecular simulation: a computational microscope for molecular biology.
- Record: found
- Abstract: found
- Article: not found
Methods for the directed evolution of proteins.
- Record: found
- Abstract: found
- Article: not found