- Record: found
- Abstract: found
- Article: found
Functions of MutLα, Replication Protein A (RPA), and HMGB1 in 5′-Directed Mismatch Repair*
Read this article at
Abstract
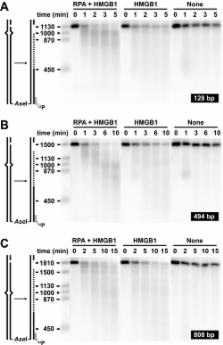
A purified system comprised of MutSα, MutLα, exonuclease 1 (Exo1), and replication protein A (RPA) (in the absence or presence of HMGB1) supports 5′-directed mismatch-provoked excision that terminates after mismatch removal. MutLα is not essential for this reaction but enhances excision termination, although the basis of this effect has been uncertain. One model attributes the primary termination function in this system to RPA, with MutLα functioning in a secondary capacity by suppressing Exo1 hydrolysis of mismatch-free DNA (Genschel, J., and Modrich, P. (2003) Mol. Cell 12, 1077–1086). A second invokes MutLα as the primary effector of excision termination (Zhang, Y., Yuan, F., Presnell, S. R., Tian, K., Gao, Y., Tomkinson, A. E., Gu, L., and Li, G. M. (2005) Cell 122, 693–705). In the latter model, RPA provides a secondary termination function, but together with HMGB1, also participates in earlier steps of the reaction. To distinguish between these models, we have reanalyzed the functions of MutLα, RPA, and HMGB1 in 5′-directed mismatch-provoked excision using purified components as well as mammalian cell extracts. Analysis of extracts derived from A2780/AD cells, which are devoid of MutLα but nevertheless support 5′-directed mismatch repair, has demonstrated that 5′-directed excision terminates normally in the absence of MutLα. Experiments using purified components confirm a primary role for RPA in terminating excision by MutSα-activated Exo1 but are inconsistent with direct participation of MutLα in this process. While HMGB1 attenuates excision by activated Exo1, this effect is distinct from that mediated by RPA. Assay of extracts derived from HMGB1 +/+ and HMGB1 −/− mouse embryo fibroblast cells indicates that HMGB1 is not essential for mismatch repair.
Related collections
Most cited references30
- Record: found
- Abstract: found
- Article: not found
Mechanisms and functions of DNA mismatch repair.
- Record: found
- Abstract: found
- Article: not found
The multifaceted mismatch-repair system.
- Record: found
- Abstract: not found
- Article: not found