- Record: found
- Abstract: found
- Article: found
Glucose repression in Saccharomyces cerevisiae

Read this article at
Abstract
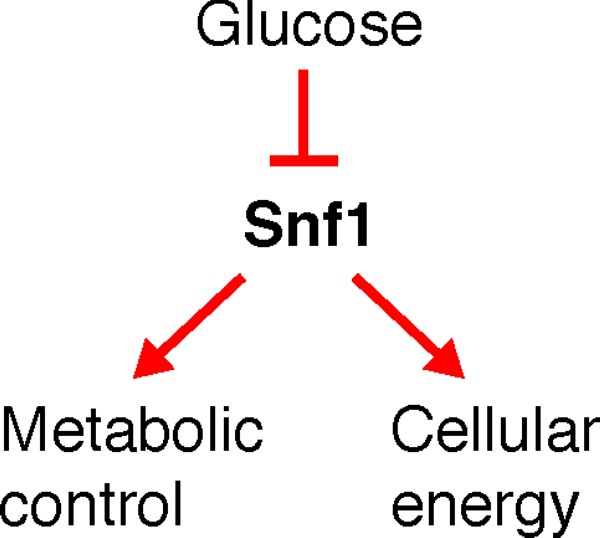
Glucose is the primary source of energy for the budding yeast Saccharomyces cerevisiae. Although yeast cells can utilize a wide range of carbon sources, presence of glucose suppresses molecular activities involved in the use of alternate carbon sources as well as it represses respiration and gluconeogenesis. This dominant effect of glucose on yeast carbon metabolism is coordinated by several signaling and metabolic interactions that mainly regulate transcriptional activity but are also effective at post-transcriptional and post-translational levels. This review describes effects of glucose repression on yeast carbon metabolism with a focus on roles of the Snf3/Rgt2 glucose-sensing pathway and Snf1 signal transduction in establishment and relief of glucose repression.
Abstract
The role of Snf1 signaling in glucose repression and carbon metabolism in Saccharomyces cerevisae.
Abstract
Related collections
Most cited references78
- Record: found
- Abstract: found
- Article: not found
Global analysis of protein phosphorylation in yeast.
- Record: found
- Abstract: found
- Article: not found
Gene expression is circular: factors for mRNA degradation also foster mRNA synthesis.
- Record: found
- Abstract: found
- Article: not found