- Record: found
- Abstract: found
- Article: found
Metabolomics study on primary dysmenorrhea patients during the luteal regression stage based on ultra performance liquid chromatography coupled with quadrupole-time-of-flight mass spectrometry
Read this article at
Abstract
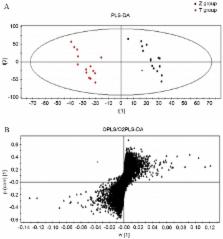
Primary dysmenorrhea (PD) is a common gynecological disorder which, while not life-threatening, severely affects the quality of life of women. Most patients with PD suffer ovarian hormone imbalances caused by uterine contraction, which results in dysmenorrhea. PD patients may also suffer from increases in estrogen levels caused by increased levels of prostaglandin synthesis and release during luteal regression and early menstruation. Although PD pathogenesis has been previously reported on, these studies only examined the menstrual period and neglected the importance of the luteal regression stage. Therefore, the present study used urine metabolomics to examine changes in endogenous substances and detect urine biomarkers for PD during luteal regression. Ultra performance liquid chromatography coupled with quadrupole-time-of-flight mass spectrometry was used to create metabolomic profiles for 36 patients with PD and 27 healthy controls. Principal component analysis and partial least squares discriminate analysis were used to investigate the metabolic alterations associated with PD. Ten biomarkers for PD were identified, including ornithine, dihydrocortisol, histidine, citrulline, sphinganine, phytosphingosine, progesterone, 17-hydroxyprogesterone, androstenedione, and 15-keto-prostaglandin F2α. The specificity and sensitivity of these biomarkers was assessed based on the area under the curve of receiver operator characteristic curves, which can be used to distinguish patients with PD from healthy controls. These results provide novel targets for the treatment of PD.
Related collections
Most cited references34

- Record: found
- Abstract: found
- Article: found
HMDB: a knowledgebase for the human metabolome
- Record: found
- Abstract: found
- Article: not found
MetPA: a web-based metabolomics tool for pathway analysis and visualization.
- Record: found
- Abstract: not found
- Article: not found