- Record: found
- Abstract: found
- Article: found
Identification of Important Modules and Hub Gene in Chronic Kidney Disease Based on WGCNA
Read this article at
Abstract
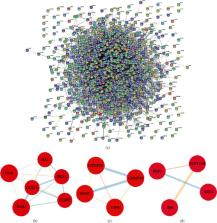
Chronic kidney disease (CKD) is an ongoing deterioration of renal function that often progresses to end-stage renal disease. In this study, we aimed to screen and identify potential key genes for CKD using the weighted gene coexpression network (WGCNA) analysis tool. Gene expression data related to CKD were screened from GEO database, and expression datasets of GSE66494 and GSE62792 were obtained. After discrete analysis of samples, WGCNA analysis was performed to construct gene coexpression module, and the correlation between the module and disease was calculated. The modules with a significant correlation with the disease were selected for Gene Ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis. Then, the interaction network of related molecules was constructed, and the high score subnetwork was selected, and the candidate key molecules were identified. A total of 882 DEGs were identified in the screening datasets. A subnetwork containing 6 nodes was found with a high score of 12.08, including CEBPZ, IFI16, LYAR, BRIX1, BMS1, and DDX18. DEGs could significantly differentiate CKD and healthy individuals in principal component analysis. In addition, the MEturquiose, MEred, and MEblue in group were significantly correlated with disease in WGCNA. These 6 hub genes were found to significantly discriminate between CKD and healthy controls in the validation dataset, suggesting that they could use these molecules as candidate markers to distinguish CKD from healthy people. Overall, our study indicated that 6 hub genes may play key roles in the occurrence and development of CKD.
Related collections
Most cited references26

- Record: found
- Abstract: found
- Article: found
WGCNA: an R package for weighted correlation network analysis

- Record: found
- Abstract: found
- Article: found
NCBI GEO: archive for functional genomics data sets—update
- Record: found
- Abstract: not found
- Article: not found