- Record: found
- Abstract: found
- Article: found
Rhamnolipid Enhances the Nitrogen Fixation Activity of Azotobacter chroococcum by Influencing Lysine Succinylation
Read this article at
Abstract
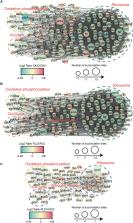
The enhancement of nitrogen fixation activity of diazotrophs is essential for safe crop production. Lysine succinylation (K Suc) is widely present in eukaryotes and prokaryotes and regulates various biological process. However, knowledge of the extent of K Suc in nitrogen fixation of Azotobacter chroococcum is scarce. In this study, we found that 250 mg/l of rhamnolipid (RL) significantly increased the nitrogen fixation activity of A. chroococcum by 39%, as compared with the control. Real-time quantitative reverse transcription PCR (qRT-PCR) confirmed that RL could remarkably increase the transcript levels of nifA and nifHDK genes. In addition, a global K Suc of A. chroococcum was profiled using a 4D label-free quantitative proteomic approach. In total, 5,008 K Suc sites were identified on 1,376 succinylated proteins. Bioinformatics analysis showed that the addition of RL influence on the K Suc level, and the succinylated proteins were involved in various metabolic processes, particularly enriched in oxidative phosphorylation, tricarboxylic acid cycle (TCA) cycle, and nitrogen metabolism. Meanwhile, multiple succinylation sites on MoFe protein (NifDK) may influence nitrogenase activity. These results would provide an experimental basis for the regulation of biological nitrogen fixation with K Suc and shed new light on the mechanistic study of nitrogen fixation.
Related collections
Most cited references62
- Record: found
- Abstract: found
- Article: not found
Cytoscape: a software environment for integrated models of biomolecular interaction networks.
- Record: found
- Abstract: found
- Article: not found
MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification.

- Record: found
- Abstract: found
- Article: found
