- Record: found
- Abstract: found
- Article: found
Reversible Dual-Covalent Molecular Locking of the 14-3-3/ERRγ Protein–Protein Interaction as a Molecular Glue Drug Discovery Approach
Read this article at
Abstract
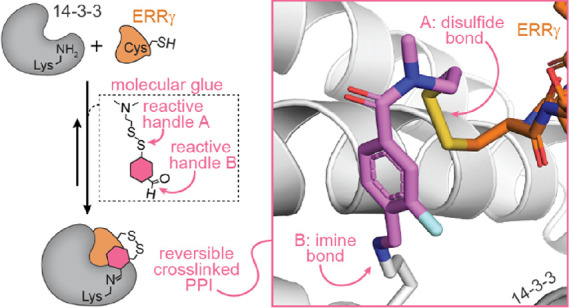
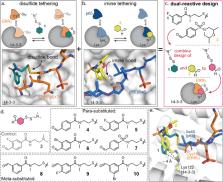
Molecules that stabilize protein–protein interactions (PPIs) are invaluable as tool compounds for biophysics and (structural) biology, and as starting points for molecular glue drug discovery. However, identifying initial starting points for PPI stabilizing matter is highly challenging, and chemical optimization is labor-intensive. Inspired by chemical crosslinking and reversible covalent fragment-based drug discovery, we developed an approach that we term “molecular locks” to rapidly access molecular glue-like tool compounds. These dual-covalent small molecules reversibly react with a nucleophilic amino acid on each of the partner proteins to dynamically crosslink the protein complex. The PPI between the hub protein 14-3-3 and estrogen-related receptor γ (ERRγ) was used as a pharmacologically relevant case study. Based on a focused library of dual-reactive small molecules, a molecular glue tool compound was rapidly developed. Biochemical assays and X-ray crystallographic studies validated the ternary covalent complex formation and overall PPI stabilization via dynamic covalent crosslinking. The molecular lock approach is highly selective for the specific 14-3-3/ERRγ complex, over other 14-3-3 complexes. This selectivity is driven by the interplay of molecular reactivity and molecular recognition of the composite PPI binding interface. The long lifetime of the dual-covalent locks enabled the selective stabilization of the 14-3-3/ERRγ complex even in the presence of several other competing 14-3-3 clients with higher intrinsic binding affinities. The molecular lock approach enables systematic, selective, and potent stabilization of protein complexes to support molecular glue drug discovery.
Related collections
Most cited references55
- Record: found
- Abstract: found
- Article: not found
Lenalidomide induces ubiquitination and degradation of CK1α in del(5q) MDS
- Record: found
- Abstract: found
- Article: found
α-Ketoamides as Broad-Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, Synthesis, and Activity Assessment
- Record: found
- Abstract: found
- Article: not found