- Record: found
- Abstract: found
- Article: found
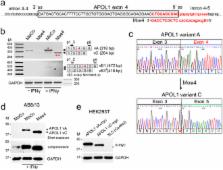
Blocking the 5′ splice site of exon 4 by a morpholino oligomer triggers APOL1 protein isoform switch
Read this article at
Abstract
APOL1 risk alleles G1 or G2 are associated with a kidney disease phenotype exclusively in people of recent African ancestry. Here we show that exon 4 encoding a part of the APOL1 signal peptide is constitutively spliced in major APOL1 transcripts expressed in kidney glomerular and tubular cells. We demonstrate that constitutive splicing of exon 4 results from a suboptimal hnRNP A1 binding motif found in exon 4. Accordingly, a robust binding of hnRNP A1 protein to a consensus hnRNP A1 cis-acting element in exon 4 results in almost complete exclusion of exon 4 from the APOL1 minigene transcripts. Blocking the 5′ splice site at the exon 4/intron boundary with a specific antisense morpholino oligonucleotide excludes exon 4 from the splicing pattern of endogenous APOL1 transcripts. These transcripts are fully functional and produce APOL1 protein isoform that is not normally detectable in podocytes. Together with our previous data showing no cytotoxicity of overexpressed APOL1 isoform lacking exon 4, we propose that morpholino-induced APOL1 isoform switch may provide a new tool to identify in vivo molecular mechanism(s) by which risk alleles promote or mediate the kidney disease phenotype.
Related collections
Most cited references32
- Record: found
- Abstract: found
- Article: not found
Splicing regulation: from a parts list of regulatory elements to an integrated splicing code.
- Record: found
- Abstract: found
- Article: not found
APOL1 risk variants, race, and progression of chronic kidney disease.
- Record: found
- Abstract: found
- Article: not found