- Record: found
- Abstract: found
- Article: found
The Fanconi Anemia DNA Repair Pathway Is Regulated by an Interaction between Ubiquitin and the E2-like Fold Domain of FANCL*
Read this article at
Abstract
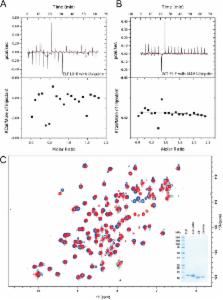
Background: The E3 ligase FANCL monoubiquitinates FANCD2 in a critical step in the repair of DNA interstrand crosslinks.
Results: FANCL binds ubiquitin non-covalently via its N-terminal E2-like fold.
Conclusion: Monoubiquitination of FANCD2 is regulated via a non-covalent interaction between FANCL and ubiquitin.
Significance: This interaction represents an additional layer of regulation of the Fanconi Anemia pathway, and a targetable interface.
Abstract
The Fanconi Anemia (FA) DNA repair pathway is essential for the recognition and repair of DNA interstrand crosslinks (ICL). Inefficient repair of these ICL can lead to leukemia and bone marrow failure. A critical step in the pathway is the monoubiquitination of FANCD2 by the RING E3 ligase FANCL. FANCL comprises 3 domains, a RING domain that interacts with E2 conjugating enzymes, a central domain required for substrate interaction, and an N-terminal E2-like fold (ELF) domain. The ELF domain is found in all FANCL homologues, yet the function of the domain remains unknown. We report here that the ELF domain of FANCL is required to mediate a non-covalent interaction between FANCL and ubiquitin. The interaction involves the canonical Ile44 patch on ubiquitin, and a functionally conserved patch on FANCL. We show that the interaction is not necessary for the recognition of the core complex, it does not enhance the interaction between FANCL and Ube2T, and is not required for FANCD2 monoubiquitination in vitro. However, we demonstrate that the ELF domain is required to promote efficient DNA damage-induced FANCD2 monoubiquitination in vertebrate cells, suggesting an important function of ubiquitin binding by FANCL in vivo.
Related collections
Most cited references38
- Record: found
- Abstract: found
- Article: not found
RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO.
- Record: found
- Abstract: found
- Article: not found
Ubiquitin-binding domains - from structures to functions.
- Record: found
- Abstract: found
- Article: not found